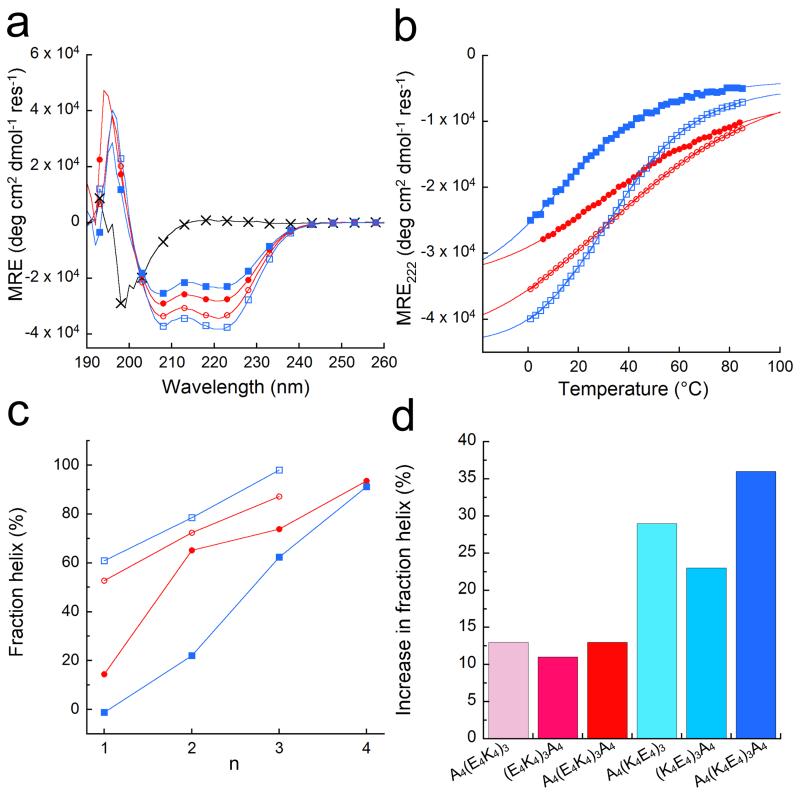
Figure 2. Helicities of the designed peptides in solution.
(a) CD spectra at 5 °C of: (E2K2)6 (black crosses), (E4K4)3 (red circles), (K4E4)3 (blue squares), A4(E4K4)3A4 (red open circles), and A4(K4E4)3A4 (blue open squares). (b) Thermal unfolding curves followed by the change in MRE222 for all except (E2K2)6, with the symbols and colours from (a) preserved. Fits to the Gibbs-Helmholtz equation are shown by solid lines. (c) Fraction helix as a function of peptide length for the (E4K4)n-(red) and (K4E4)n-based (blue) peptides with (open symbols) and without (filled symbols) tetra-alanine flanks (data points are joined by lines to guide the eye). (d) Increase in fraction helicities at 5 °C of the alanine-flanked peptides A4(E4K4)3, (E4K4)3A4 and A4(E4K4)3A4 (red colours) relative to (E4K4)3; and for A4(K4E4)3, (K4E4)3A4 and A4(K4E4)3A4 (blue colours) relative to (K4E4)3. Conditions: 100 μM peptide concentration in PBS 137 mM NaCl, pH 7.4.