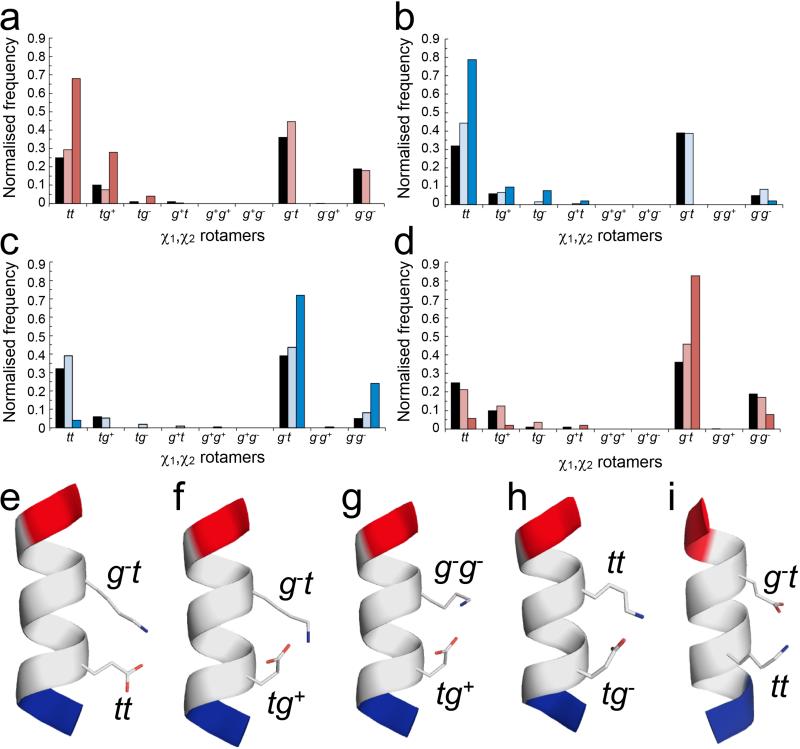
Figure 4. Side-chain interactions observed in α-helices from the Protein Data Bank.
χ1,χ2 distributions and conformers for Ei➔Ki+4 and Ki➔Ei+4 pairs in high-resolution X-ray crystal structures. (a – d) Normalised frequencies of preferred χ1,χ2 angles for Glu (a&d) and Lys (b&c) residues in Ei➔Ki+4 (a&c) and Ki➔Ei+4 (b&d) pairs. Key: red for Glu, blue for Lys; black bars indicate the frequency of each rotamer found in all α-helices; pale bars indicate pairs where no salt bridge is made; and dark bars indicate pairs where a salt bridge is formed. (e – i) Examples of salt-bridging pairs. For the Ei➔Ki+4 pairs, there are two dominant rotamer combinations, with Glu (tt; tg+) plus Lys (g−t) (e&f, 17 examples (68%) and 6 examples (24%), respectively), and two minor combinations (g&h, 1 example of each (8%)) with Glu;Lys (tg+;g−g−) and Glu;Lys (tg−; tt). Whereas for the Ki➔Ei+4 pairs there is one clearly preferred conformation (Glu, g−t; Lys, tt) (i, 39 examples (75%)). Only the g−t;g−t combination would be better, but inspection of molecular models revealed that with χ1 = g− for Lys takes the ε-amino group too far from the γ-carboxylate of Glu to form a salt bridge. Examples were taken from the PDB as follows: (e) 1kqp A246-A250; (f) 1moq A481-A485; (g) 3n0u A177-A181; (h) 2r75 A141-A145; (i) 1egw A30-A34. Images were generated with PyMol (www.pymol.org).