Fig. 3.
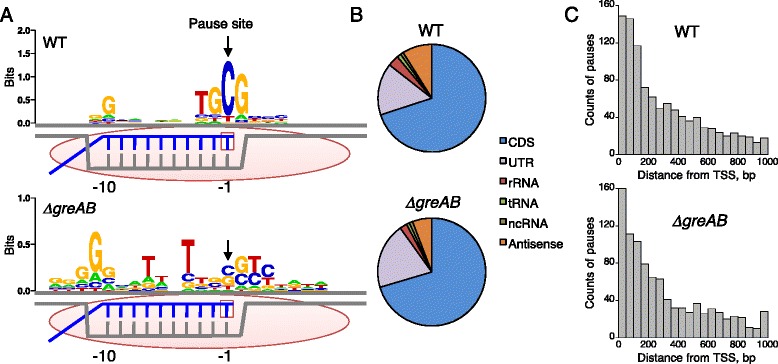
Transcription pausing detected by RNET-seq in E. coli WT and ΔgreAB cells. (a) Pause-inducing elements (PIEs) of the non-template DNA strand. Information content is represented by sequence logos [51]. Positions −1 and −10 of DNA (gray) correspond to the RNA (blue) 3′ and 5′ ends of the RNA-DNA hybrid within RNAP (pink oval) in pre-translocated ECs. The active site is shown as an empty square. P(0.9, 100) and P(0.9, 160) were used for WT (n = 758) and ΔgreAB (n =419), respectively (see main text for the parameters). A frequency matrix and MAP scores for the PIEs are shown in Table S1 in Additional file 2. (b) Categorization of all RNAP pauses by RNA type. The non-coding RNA (ncRNA) and antisense RNA were defined using the gene annotation file of E. coli (see “Materials and methods”). (c) Pausing frequently occurs in regions proximal to transcript start sites (TSS). For panels B and C, P(0.7, 100) is used in order to increase the number of samples. Note that even when using this reduced stringency the consensus sequence for pausing remains unaffected (Fig. S6B in Additional file 1)
