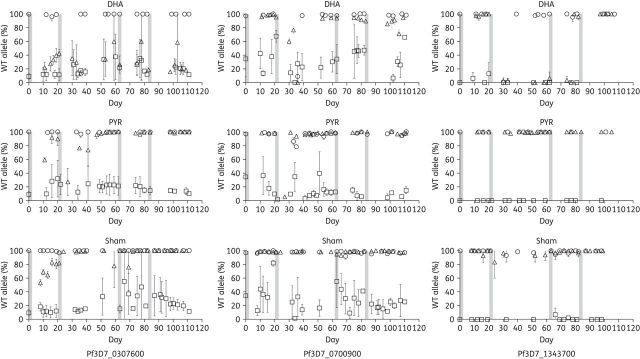
Figure 4.
Artemisinin-resistant allele frequency. Allele frequency of SNPs was assessed by pyrosequence analysis and is plotted as the percentage of WT (D6) SNP found at each timepoint in the competitive growth experiment. Drug or sham exposures were for 48 h and are denoted as bars. The SNPs quantified were on chromosomes 3 (Pf3D7_0307600; conserved protein with unknown function), 7 (Pf3D7_0700900; RESA-like protein, pseudogene) and 13 (Pf3D7_1343700, K13). The Pf3D7_0700900 and Pf3D7_1343700 SNPs were lost in C9 and the mixed culture in the absence of dihydroartemisinin pressure. Conversely, the Pf3D7_0307600 SNP persisted throughout the study in C9 and the mixed culture exposed to dihydroartemisinin. The K13 SNP was maintained in artemisinin-resistant C9 over four consecutive treatments plotted as proportion of WT allele. DHA, dihydroartemisinin; PYR, pyrimethamine.