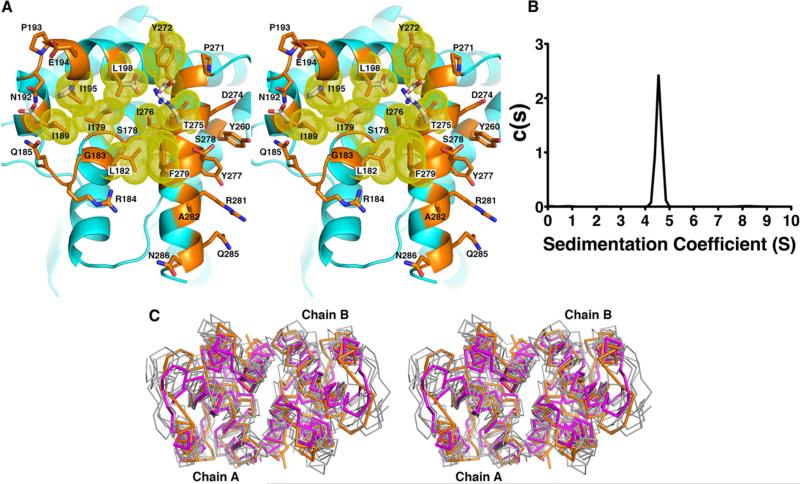
Figure 4.
KPR dimer interface. (A) The dimer interface is composed of a hydrophobic pocket. Residues within the pocket are colored orange with yellow van der Waals radii and are listed in Table 2. White side chains are polar residues found in the E. coli KPR. (B) Analytical ultracentrifugation experiments showed only one species of KPR in solution at 4.5 S, corresponding to the dimer as predicted by HYDROPRO.18 (C) Stereoview of the superposition of the Cα traces of the C-terminal domains of six homologous (unpublished) KPR dimers (gray) onto S. aureus KPR (orange). The related enzyme, d-mandelate dehydrogenase of Enterococcus faecium31 (magenta), is also shown. Enzymes are from Ralstonia eutropha (PDB entry 3HWR, 20% sequence identity), Ralstonia solanacearum (PDB entry 3GHY, 21% sequence identity), Mycobacterium tuberculosis (PDB entry 4OL9, 31% sequence identity), Enterococcus faecalis (PDB entry 2EW2, 20% sequence identity), Methylococcus capsulatus (PDB entry 3I83, 20% sequence identity), Bacillus subtilis (PDB entry 3EGO, 25% sequence identity), and Ent. faecium (PDB entry 3WFJ, 21% sequence identity).