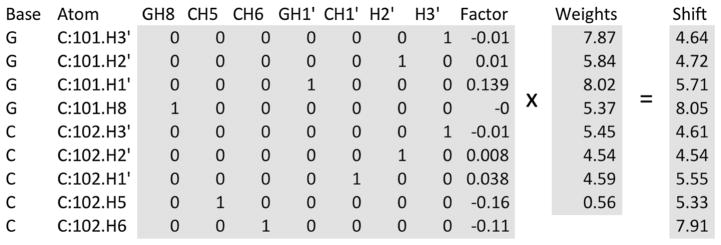
Fig. 2.
A representation of the math matrices used for calibrating ring-current shift parameters. The first and last shaded regions represent the matrix A and vector b that are input into the Singular Value Decomposition (SVD) algorithm to solve the equation Ax = b for the vector x (representing the contribution of each term). The actual matrix A used has additional rows for the remaining atoms in the structure, and additional columns for additional atom types (for the A and U residues), and the vector b has additional rows (for the additional atoms). The final column, labeled Factor, represents the sum of product of the geometric factors and ring current intensity parameter (Case 1995) calculated for all aromatic rings near the corresponding atom. All the other columns of the matrix are set to 0, except the column that represents the atom type for that row