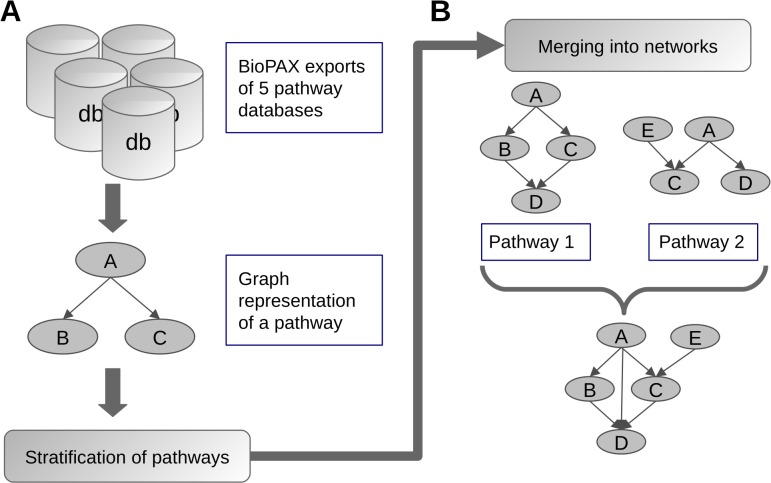
Fig 1. Workflow of parsing and merging pathways.
BioPAX encoded pathway data on WNT signaling was collected across 5 databases, parsed into R, and transformed into signaling graphs (A). The pathways were stratified into conceptual groups and within each group the graphs were sequentially merged into a signaling network. When merging two pathways, the resulting graph contains all nodes and edges from both original graphs (B).