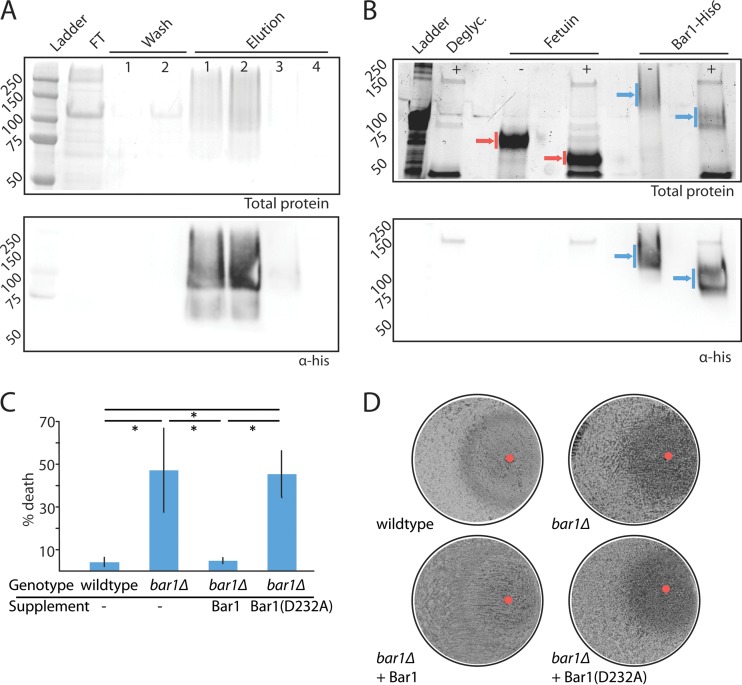
FIG 1 .
Purification and analysis of C. albicans Bar1 protease. (A) Purification of His-tagged C. albicans Bar1 from Pichia pastoris using nickel affinity chromatography. Nickel beads were washed twice (20 mM imidazole), and protein was eluted with 500 mM imidazole. FT, flowthrough. Total protein was assayed by Coomassie staining (top panel), and Bar1 was detected by Western blotting with an anti-His antibody (lower panel). (B) Protein glycosylation was assessed by treatment of Bar1 or fetuin (positive control) with a cocktail of deglycosylation (Deglyc.) enzymes. Blue arrows, Bar1; red arrows, fetuin. The presence or absence of deglycosylase enzymes is noted with ±. Total protein is shown (top panel) using BioRad Stainfree indicator, and Bar1 was detected by Western blotting with an anti-His antibody (lower panel). (C) To evaluate the activity of recombinant Bar1, wild-type and bar1Δ/bar1Δ opaque a strains were treated with or without 300 nM α pheromone for 5 h, together with recombinant Bar1 or Bar1(D232A) at 127 nM. The percentage of death was analyzed by staining with propidium iodide and flow cytometry. Values are means ± standard deviations (SD). n = 3. *, P < 0.05 by t test. (D) Cell cycle arrest was analyzed by plating a lawn of opaque a cells on solid medium and spotting 20 µg α pheromone (red circle) together with Bar1 or Bar1(D232A), followed by imaging of plates after 3 days of culture at 22°C.