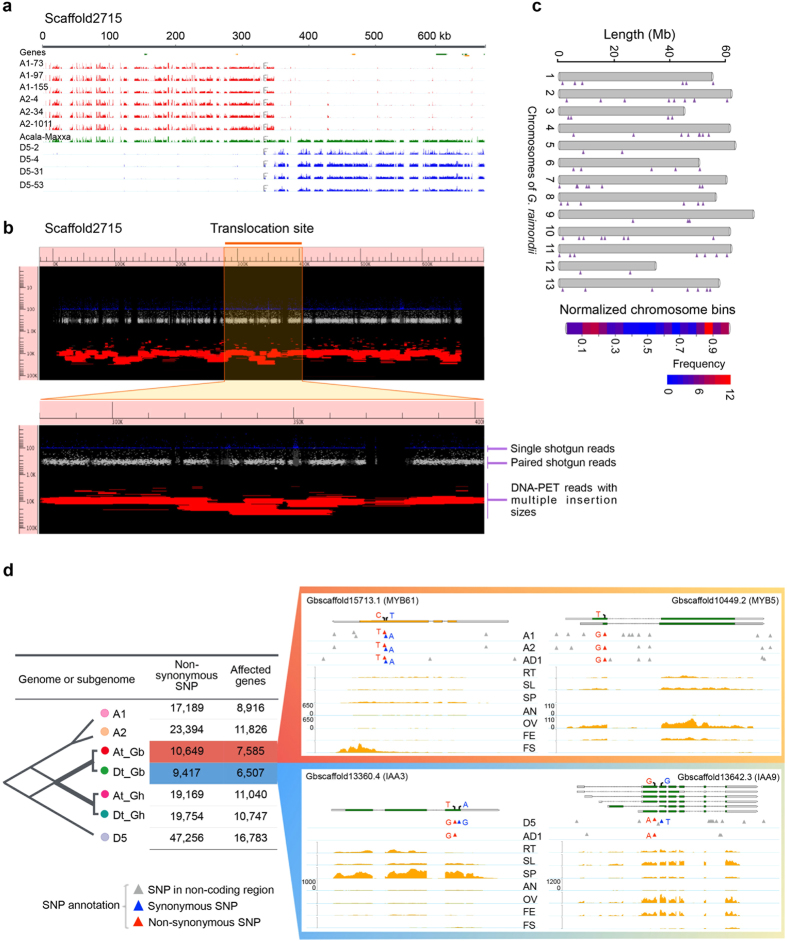
Figure 3. Impact of allotetraploidization on genomic variation.
(a) An example of At-Dt-hybrid scaffold. The mapping tracks, order and density scale are same as in Fig. 1b. (b) Evaluating assembly quality of hybrid scaffold at junction region. The y-axis indicates that the fragment size of paired-end reads in log scale, the x-axis indicates the length of the scaffold. As shown, single-end shotgun reads (blue, ~100 bp) and paired-end shotgun reads (white, ~300 bp) were clustered into contigs, which were further connected by abundant DNA-PET reads (size range of 5, 10, and 20 Kb) in this scaffold. (c) Genome-wide distribution of inter-subgenome translocations. Using the D5 genome reference as a framework, all 77 putative translocation sites are indicated along the 13 chromosomes by triangles. (d) Lineage-specific SNP divergence in diploid genomes and tetraploid subgenomes. The right panel provides examples of genes that were affected by lineage-specific non-synonymous SNPs in subgenomes of G. barbadense (two MYB genes holding lineage-specific SNPs in At and two auxin-responsive factor genes holding lineage-specific SNPs in Dt). The RNA-Seq tracks show the expression profile of the four genes (the details of sample was listed in the Supplementary Table 10) and adjusted on the same scale.