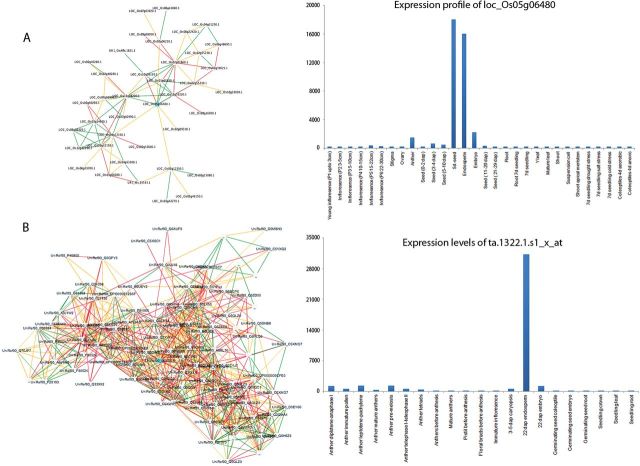
Fig. 4.
The gene regulatory networks derived from co-expression data of seed development-specific genes. (A) The gene regulatory networks of the endosperm-specific H + -translocating pyrophosphatase gene cloned for chalkiness in rice derived using the PLANET database (Mutwil et al., 2011) seem to control the selective proteolysis pathway (E3.SCF) during 3–5 DAF (Supplementary Table S2 at JXB online). (B) On the other hand, its orthologue in wheat (Ta.1322.1.S1_x_at) regulates thousands of genes involved in starch biosynthesis and protease inhibitors during seed filling. These divergent expression patterns of H + -translocating pyrophosphatase genes between rice and triticale members are likely to trigger functional divergence during seed development.