Fig. 6.
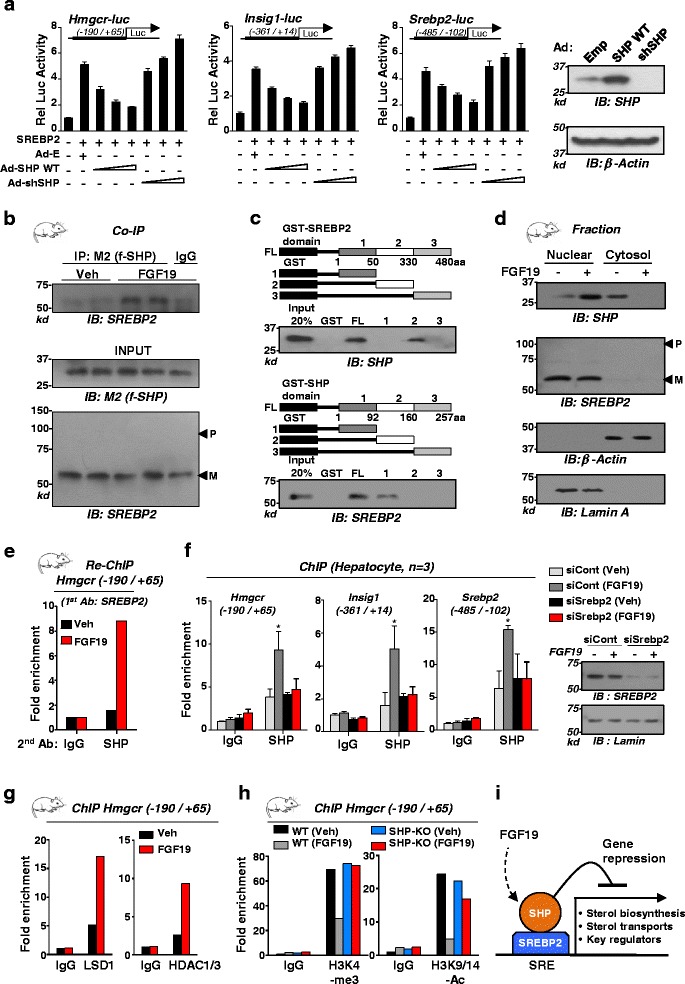
FGF19 increased functional interaction of SHP with SREBP-2, resulting in epigenomic repression. a Reporter assay: HepG2 cells were transfected with plasmids and infected with adenoviruses as indicated, treated with FGF19, and luciferase activity was normalized to β-galactosidase activity. Protein levels of SHP are shown at right. b CoIP: Mice were tail vein injected with Ad-flag-SHP, 1 week after infection, mice were treated with vehicle or FGF19 for 2 h, and CoIP assays using liver nuclear extracts were performed. P or M indicates the precursor or mature form of SREBP-2, respectively. c GST pull-down: Schematics of the SHP and SREBP-2 domains fused to GST are shown at the top. The amount of SHP and SREBP-2 bound to the reciprocal GST-proteins were determined by IB. d Fractionation study: Effects of FGF19 on protein levels of SHP and SREBP-2 in nuclear and cytoplasmic fractions of mouse liver extracts. Actin and lamin were markers for the cytoplasm and nucleus, respectively. e Liver re-ChIP: Mice were treated with vehicle or FGF19 for 2 h and livers were pooled from three mice. Chromatin was immunoprecipitated with SREBP-2 antibody first and then eluted and re-precipitated with SHP antibody. f siRNA/ChIP: Hepatocytes were transfected with siRNA for SREBP-2 or control RNA and then 3 days later, cells were treated with vehicle or FGF19 for 2 h and ChIP assays were done. Statistical significance was determined by the Student’s t-test (SEM, n = 3, *P <0.05). At right, levels of nuclear SREBP-2 protein levels detected by IB are shown. g, h ChIP: WT or SHP-KO mice (pooled from three mice) were treated with vehicle or FGF19 and ChIP assays were done. i Model illustrating molecular mechanisms by which SHP inhibits SREBP-2 direct target genes
