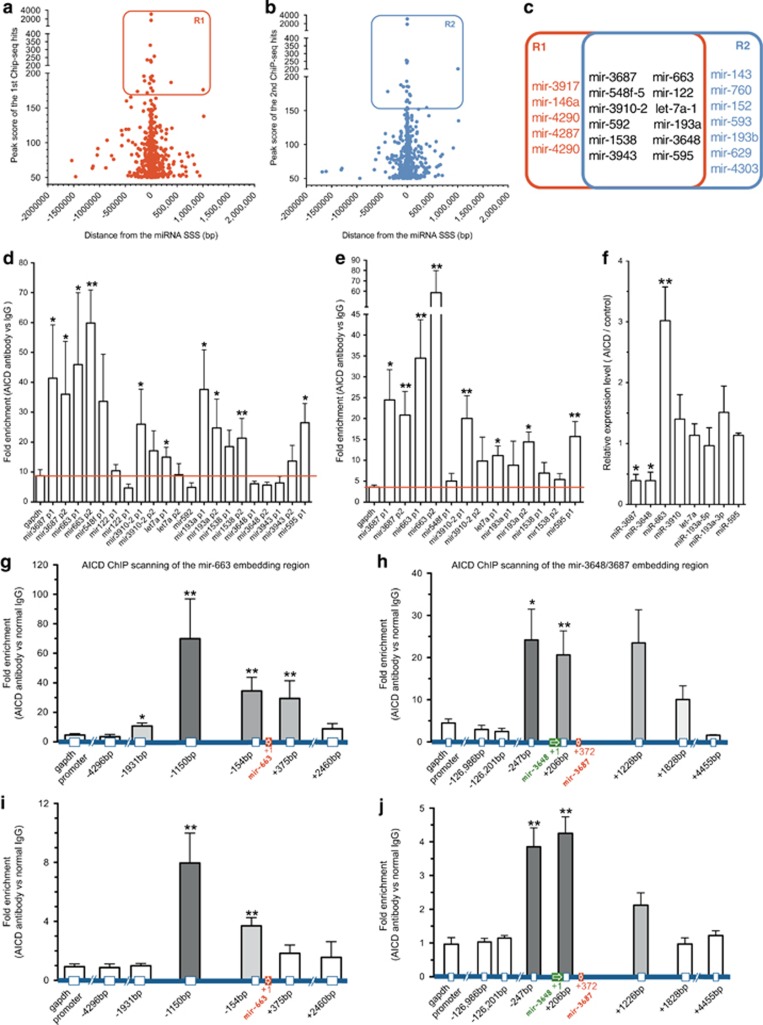
Figure 2.
ChIP validation and miRNA expression assay for possible miRNAs regulated by AICD in hNSCs. (a and b) Top 20 'peak score' miRNA hits were selected from the first (R1; a) and the second ChIP-seq data set (R2; b). (c) The overlapping 12 miRNA hits from R1 and R2 were selected for ChIP validation. (d) AICD ChIP validation performed in SH-SY5Y cells overexpressing AICD. The gapdh promoter region is set as control. (e) AICD ChIP validation performed in hNSCs overexpressing AICD. (f) Expression fold change of the eight miRNAs in hNSCs after AICD overexpression. (g) AICD ChIP scanning of the mir-663-embedding region in hNSCs overexpressing AICD. The gapdh promoter and other regions are set as control. The x axis shows the base distance from the miRNA stem–loop start site, noted as +1. (h) AICD ChIP scanning of the mir-3648- and mir-3687-embedding regions in hNSCs overexpressing AICD. (i and j) ChIP validation of the endogenous interaction of AICD with mir-663 (i), mir-3648 and mir-3687 (j) embedding regions in wild-type hNSCs. Quantification data were analyzed from at least three independent experiments (mean±S.E.M). *P<0.05; **P<0.01