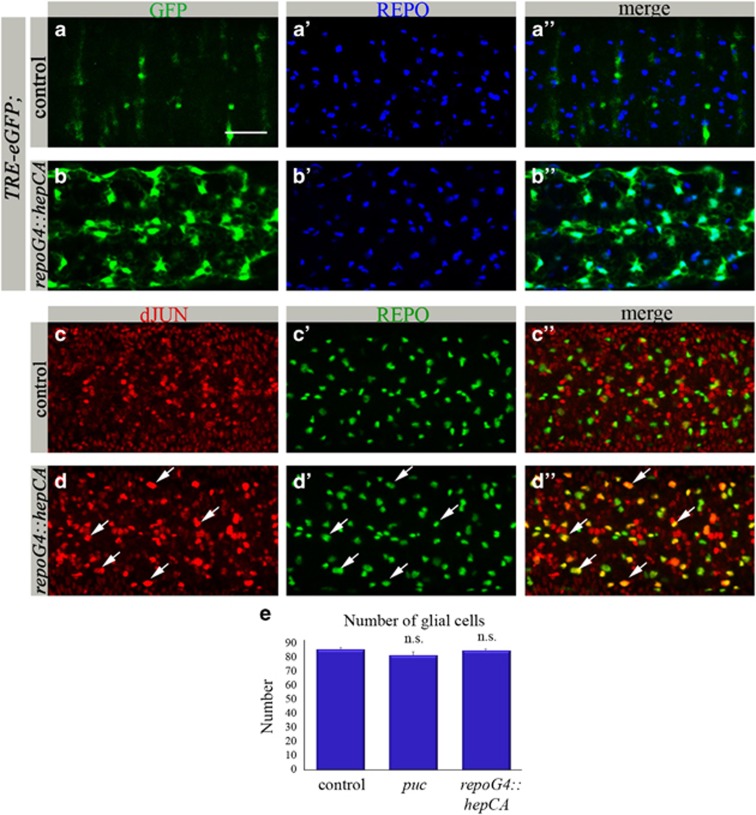
Figure 4.
Activation of dJNK signaling in glial cells promotes glial phagocytosis. (a–d″) Projections from confocal stacks of the embryonic CNS at stage 16; ventral view. Bar 20 μm. (a–b″) TRE-eGFP reporter (in green) depicts JNK pathway activation. REPO-positive glial nuclei are marked in blue. (a–a″) repoGal4 control. (b–b″) repoGal4::hepCA mutant embryo. (c–d″) anti-dJUN staining in red and anti-REPO in green. (c–c″) repoGal4 control. (d–d″) repoGal4::hepCA mutant embryo. Arrows mark glial cells expressing dJUN. (e) Number of REPO-positive nuclei within CNS sections. Columns represent mean total number of glial nuclei within confocal stacks of the CNS, ±S.E.M., n=5–8; NS (not significant) P>0.05, as determined by the Student's t-test. There is no significant difference in number of glial nuclei between repoGal4::hepCA, pucE69 mutant and control embryos