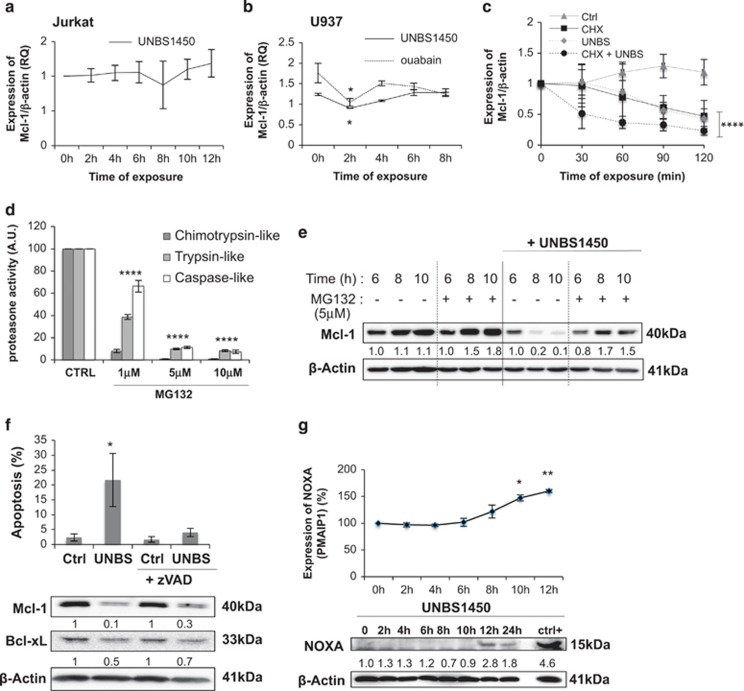
Figure 4.
UNBS1450 downregulates Mcl-1 protein expression via a mechanism involving protein stability and the proteasome. (a) After treatment of Jurkat cells with UNBS1450 (20 nM) Mcl-1 mRNA expression was measured after 0, 2, 4, 6 and 8, 10 and 12 h (a time leading to Mcl-1 protein downregulation in this cell model). (b) Same analysis was performed on U937 cells with UNBS1450 (20 nM) or ouabain (100 nM), where Mcl-1 mRNA expression was measured after 0, 2, 4, 6 and 8 h. (c) Analysis of Mcl-1 protein stability. U937 cells were incubated with 20 nM UNBS1450 and after 6 h co-treated with or without 10 μM cycloheximide (CHX) for further 2 h compared to untreated control cells (Ctrl). Samples were collected every 30 min and analyzed by western blot. Graph reports the quantification of Mcl-1 protein of 6 independent experiments +/−SD. (d) Determination of the concentration of MG132 required to inhibit the proteasome activity in U937 cells. Results are the mean of three independent experiments +/−S.D. (e) 5 μM MG132 were selected to analyze the effect on Mcl-1 protein expression in presence/absence of UNBS1450. MG132 was added after 6 h treatment with 20 nM UNBS1450. Samples were collected after 2 h for western blot analysis of Mcl-1. (f) Effects of the pan-caspase inhibitor (zVAD; 50 μM) on apoptosis (top) and Mcl-1/Bcl-xL expression (bottom) after 12 h treatment with UNBS1450. (g) Analysis of the impact of UNBS1450 on NOXA expression at mRNA and protein levels. U937 cells treated with 20-μM cisplatin for 24 h were used as positive control. Western blots are representative of three independent experiments. Results are the mean of three independent experiments +/− S.D. Statistical analysis was performed by two-way ANOVA test (post-hoc test: Dunnett or Sidak). Significance is reported as *P<0.05, **P<0.01, ****P<0.0001. In panel (b), significance is reported for UNBS1450+CHX versus CHX or UNBS1450