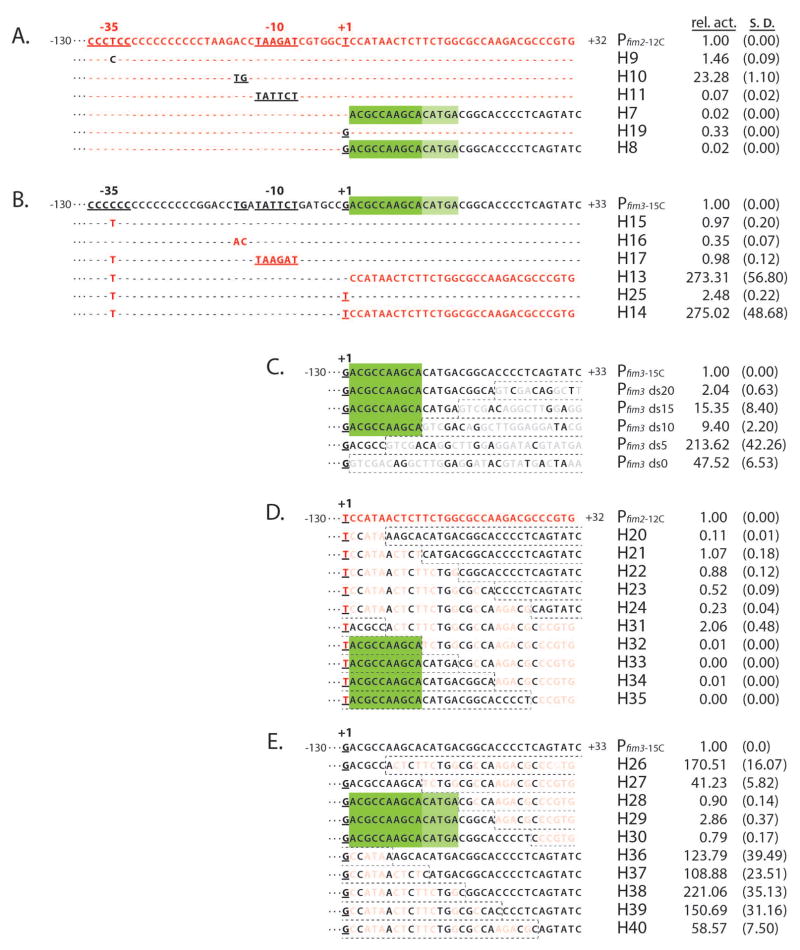
Fig. 2. DRE represses Pfim3-15C transcription in B. pertussis.
Transcriptional activities of fim hybrid promoters were assessed in vivo. Promoter fragments comprising sequences from −130 to +32 (Pfim2) or +33 (Pfim3) were cloned into the luxCDABE transcriptional vector pSS3967, the resulting constructs were inserted into the chromosome of BP536 at a constant, ectopic, location, and light output was measured as described in Experimental Procedures. Promoter activities normalized to the wild-type in each set are presented to the right, with values for standard deviation in parentheses. Based on the data in each panel, the inferred extent of the DRE is indicated by green rectangles
(A & B). Relevant portions of the nucleotide sequences of the fim2 promoter (A) and the fim3 promoter (B) are presented with sequence elements exchanged to create hybrid promoters indicated below and in contrasting color. Unchanged nucleotides are depicted by dashes.
(C). Deletion analysis of the downstream region of Pfim3. Downstream sequences were deleted to varying extents. Vector sequences substituting for promoter sequences are boxed by dashed lines. Nucleotides identical at a given position to those in the wildtype fim3 promoter are shown in black, and those that are different are depicted in light gray.
(D & E). Hybrid promoters were derived by substituting, to varying extents, sequences downstream of +1 with those of the alternate promoter. Substituting sequences are boxed by dashed lines with those differing at a given position shaded out.