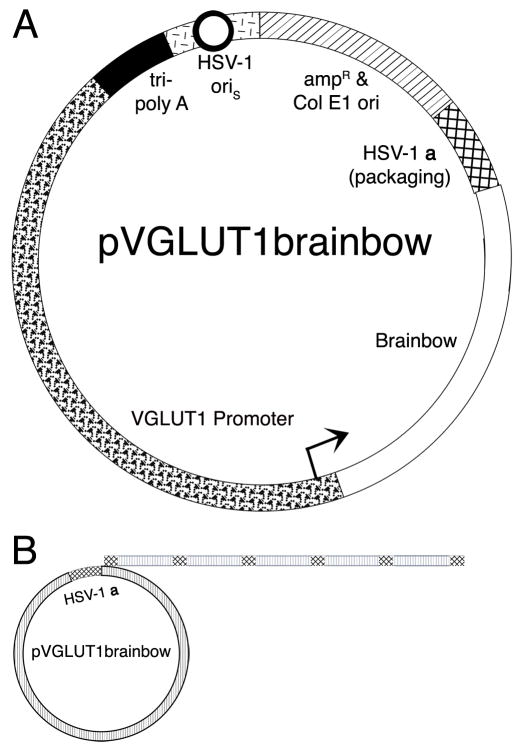
Fig. 2.
The structure of pVGLUT1brainbow and the production of Brainbow arrays by rolling circle DNA replication. (A) A schematic diagram of pVGLUT1brainbow. The VGLUT1 promoter (wavy segment with arrow) supports expression of Brainbow (clear segment). The vector backbone contains an HSV-1 origin of DNA replication (oriS, black circle with white interior in the short line segment) and an HSV-1 a sequence (cross hatched segment), which contains the packaging site; these elements support replication and packaging of the vector, respectively. A cassette of three polyadenylation sites (tri-poly A, black segment) was placed 5′ to the VGLUT1 promoter to reduce any effects on expression from the HSV-1 immediate early 4/5 promoter (contained in short line segment). Sequences from pBR322 (diagonal line segment) support growth in E. coli. (B) Concatamers of the vector are produced during the second, rolling circle phase, of HSV-1 DNA replication (Roizman and Sears, 1993). A HSV-1 genome-sized concatamer of the vector is packaged into a HSV-1 particle (Fraefel et al., 1996; Roizman and Sears, 1993); the HSV-1 genome is ~152 kb, and pVGLUT1brainbow is ~20 kb; thus, an array of 7 or 8 pVGLUT1brainbow is packaged into a HSV-1 particle.