Figure 3. Comparison of ER structures.
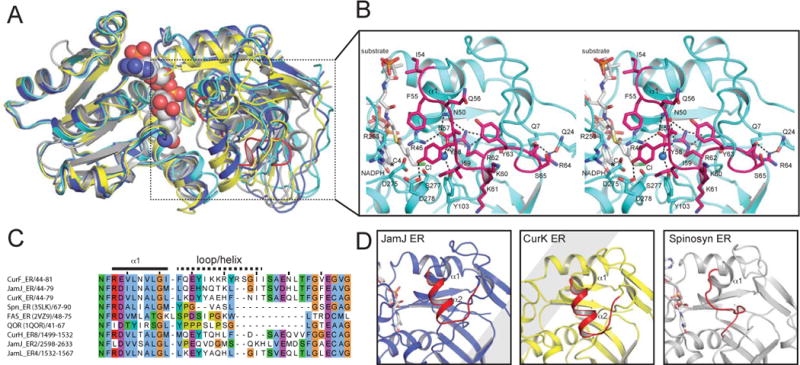
A. Superposition of nucleotide-binding domains. The similarity of nucleotide-binding domains and divergence of substrate-binding domains is apparent for NADPH-bound CurF ER (cyan), JamJ ER (blue), CurK ER (yellow) and spinosyn ER2 (grey).
B. CurF ER active site. The docked substrate (4-Cl-3-methylcrotonyl) shown in the active site in the stereo drawing (white carbon atoms). Selected residues tested by mutagenesis are shown in stick form in the cyan ER with the cyclopropanase loop highlighted in magenta. The NADPH C4 atom is marked with a star, and the active site water is rendered as a blue sphere.
C. Sequence alignment of the substrate loop region. The longer substrate loop of the cyanobacterial ERs is apparent.
D. Comparison of ER substrate loops. The longer substrate loops of the JamJ and CurK ERs include a helix, which does not exist in the shorter loop of the Spn ER2. The CurF ER cyclopropanase loop in (b) differs from the substrate loops of all canonical ERs.
