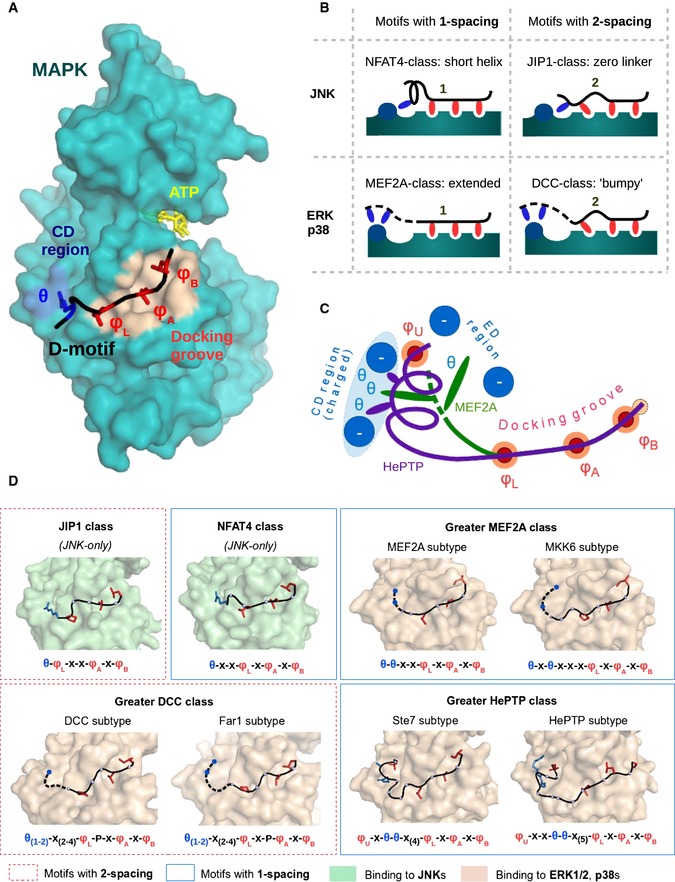
Structural heterogeneity of D‐motifs. The combinations of the three variable features yield structurally well‐defined, distinct classes of D‐motifs. Many of these models also define separate types of linear motifs, but their consensus sequences are not always exclusive. JNK kinases only admit two major types of motifs, the NFAT4 class (1‐spacing, short linker) and the JIP1 class (2‐spacing, short linker). On the other hand, known ERK1/2 and p38 binder peptides may belong to the greater MEF2A class (1‐spacing, longer linker, linear end), the greater HePTP class (1‐spacing, longer linker, helical end), or the greater DCC class (2‐spacing, longer linker, linear end). A sixth class of ERK or p38 interactors is theoretically also possible (2‐spacing, longer linker, helical end), but this combination can only be observed in long reverse (revD) motifs (Garai
et al,
2012), and no classical motif of this type is known up to date. Subtypes and other variants within a given greater class are also featured wherever applicable. These are shown based on structures of MAPK‐docking motif complexes. Dashed lines indicate N‐terminal peptide regions that are usually not visible in the crystal structures, albeit indispensable for binding. Consensus motif of each subtype is shown below, where φ
U, φ
L, φ
A, and φ
B letters denote positions that are filled by hydrophobic amino acids—L, A, and B refer to the lower pocket, and pockets A and B, respectively—while the θ positions are positively charged (Arg or Lys) while letter “x” denotes arbitrary amino acids.