Figure 2. Motif finding work flow.
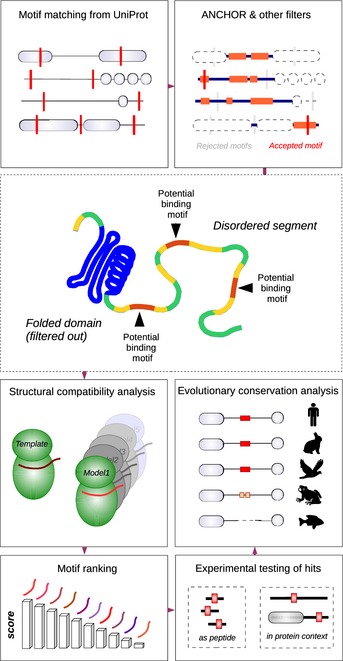
To find novel MAPK‐docking motifs, the primary motif‐matching step (on UniProt KB sequences) was followed by a series of filters. Valid motifs had to pass through an ANCHOR filter, a localization filter (combined from SignalP and Phobius) and an auxiliary Pfam filter, in order to be scored by FoldX homology models. ANCHOR (middle panel) had the most important role in selecting segments that can potentially act as linear motifs, while FoldX gave motif‐specific binding energy estimates (see Source data). Predicted motifs were subsequently tested as short fragments and (in selected cases) as full‐length proteins. Finally, an automated evolutionary analysis was performed to give information on motif conservation trends.Source data is available online for this figure.
