Figure 8. Mechanisms of binding motif emergence.
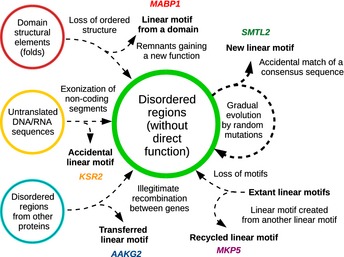
D‐motifs have diverse evolutionary origins. They most commonly originate from disordered protein segments via gradual accumulation of point mutations (e.g., SMTL2). However, they may also be created from folded domains (e.g., MABP1), or being introduced entirely de novo, from a previously untranslated genomic segment (e.g., KSR2). D‐motifs can also be born from existing linear motifs, through gene fusion or recombination (e.g., AAKG2). But more commonly, this involves transmutation of a (previous) linear motif into a new one (example: MKP5). For more details, see Table EV3 and Appendix Fig S7.
