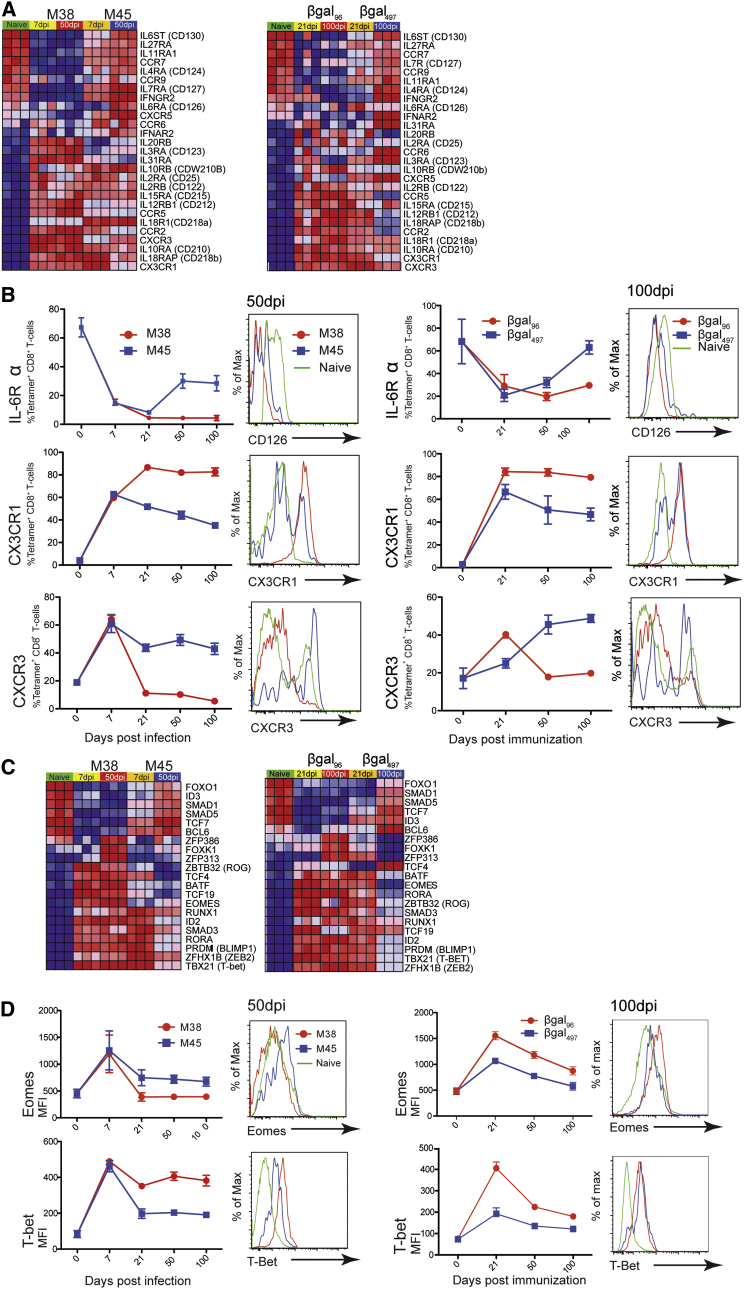
Figure 4.
Distinct Cytokine and Chemokine Receptor Expression and Individual Transcription Regulator Profile on Expanded CD8+ T Cells Compared to Conventional Memory CD8+ T Cells
(A) Heatmap showing mRNA expression levels of cytokine and chemokine receptors measured by microarray analysis of naive, M38-, and M45-specific CD8+ T cells days 7 and 50 postinfection (left panel) and naive, βgal96-, and βgal497-specific CD8+ T cells days 21 and 100 postvaccination (right panel). Shades of red indicate upregulated genes, and shades of blue indicate downregulated genes. Filter criteria of at least 2-fold changes with p ≤ 0.05, compared to naive CD8+ T cells, are shown.
(B) Longitudinal flow cytometry analysis showing the percentage of IL-6Rα (CD126), CX3CR1, and CXCR3 expression on M38/βgal96- (red) and M45/βgal497-specific CD8+ T cells (blue) in the spleen at days 0, 7, 21, 50, and 100 postinfection/immunization (n = 6–8; ±SEM). Histograms show expression of indicated markers, gated on live naive (green) M38/βgal96- (red) and M45/βgal497-specific CD8+ T cells (blue).
(C) Heatmap showing mRNA expression levels of transcriptional regulators measured by microarray analysis of naive, M38-, and M45-specific CD8+ T cells days 7 and 50 postinfection (left panel) and naive CD8+ T cells and βgal96- and βgal497-specific CD8+ T cells days 21 and 100 postvaccination (right panel). Shades of red indicate upregulated genes, and shades of blue indicate downregulated genes. Filter criteria of at least 2-fold changes with p ≤ 0.05, compared to naive CD8+ T cells, are shown.
(D) Flow cytometry analysis showing the MFI of cells expressing the TFs EOMES and Tbet on M38- (red) and M45-specific CD8+ T cells (blue; n = 6–8; ±SEM) and on βgal96- (red) and βgal497-specific (blue) CD8+ T cells in the spleen (n = 5–9; ±SEM). Histograms show expression of EOMES and Tbet and are gated on live naive (green), M38/βgal96-specific CD8+ T cells (red), and M45/βgal497-specific CD8+ T cells (blue).
See also Figures S3 and S4.