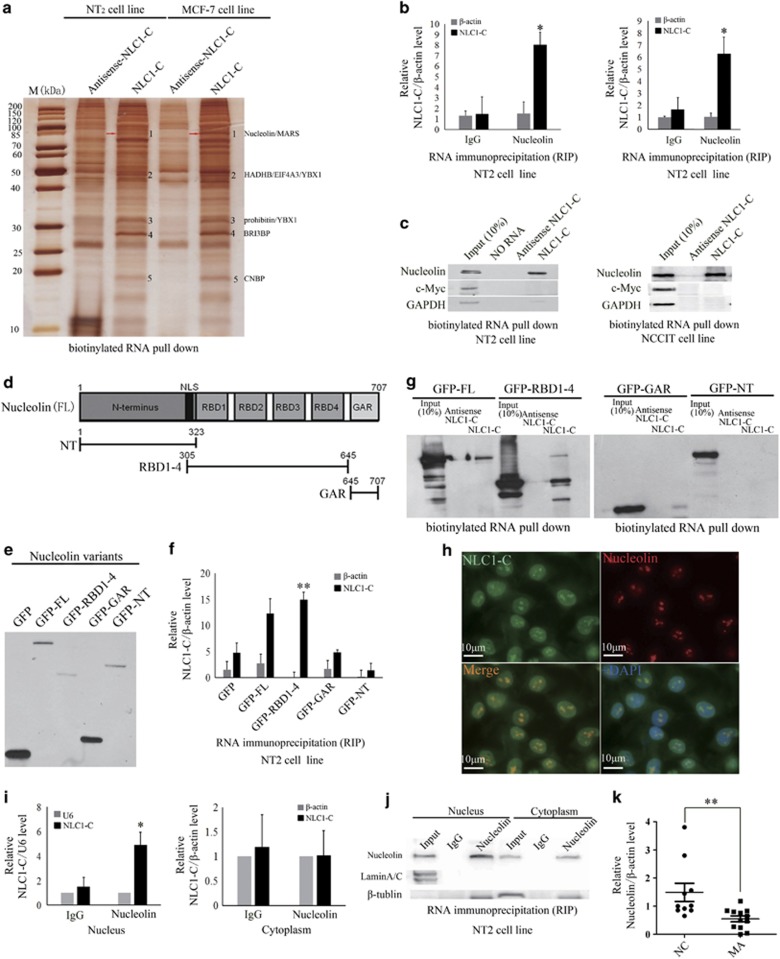
Figure 2.
NLC1-C directly binds to Nucleolin RBD domain in the nucleus. (a) Identification of cellular proteins associated with NLC1-C in vitro. Proteins from NT2 cell extracts were pulled down with the biotinylated RNAs, subjected to SDS-PAGE, and visualised by silver staining. The bands specific to NLC1-C, as indicated by the numbers (bands 1–5), were subjected to mass spectrometry. The Nucleolin (b and 1) is indicated by the red arrow. (b) NLC1-C interacts with Nucleolin in vivo. Nucleolin was immunoprecipitated from NT2 cells, NCCIT cells and co-precipitated RNAs were detected by qRT-PCR using primers for NLC1-C and β-actin (as negative control). IP enrichment was determined as the amount of RNA associated to the Nucleolin IP relative to IgG control. Data are from one of three independent experiments and are represented as mean±S.D. with n=3. (c) Immunoblotting analysis of the specific interaction of NLC1-C with Nucleolin. Proteins from NT2 and NCCIT cells pulled down with the biotinylated RNAs as in (a) were analysed by immunoblotting with a Nucleolin antibody. c-Myc and GAPDH were used as negative controls. (d) Schematic representation of the primary sequence of Nucleolin. Acidic stretches are in the N-terminal region; four RNA-binding domains (RBD), called RNA recognition motifs (RRM), are in the central region; and a glycine/arginine-rich domain or GAR domain is at the C-terminus. (e) Western blot analysis of the expression of the GFP-tagged Nucleolin constructs. The various Nucleolin constructs, as indicated, were transfected into NT2 cells for 48 h. Lysates were prepared from these cells and examined by GFP antibodies. (f) Elucidation of a direct Nucleolin–NLC1-C interaction to the RNA-binding domain. The various Nucleolin constructs, as indicated, were transfected into NT2 cells for 48 h. Lysates were prepared from these cells. RNAs that were co-precipitated with GFP were detected by qRT-PCR using primers for NLC1-C and β-actin (as negative control). IP enrichment was determined as the amount of RNA associated to the various Nucleolin domains. Data are from one of three independent experiments and are represented as mean±S.D. with n=3. (g) Immunoblotting analysis of the specific interaction of NLC1-C with the Nucleolin RBD domain. Immunoblotting analysis of the specific interaction of NLC1-C with Nucleolin. Proteins pulled down with the biotinylated RNAs as in (a) were analysed by immunoblotting with GFP antibody. (h) Co-localisation analysis: RNA FISH assay of NLC1-C combined with immunofluorescence detection of Nucleolin in NT2 cell. Scale bars, 10 μm. DAPI, 4′,6-diamidino-2-phenylindole. (i) NLC1-C directly binds to Nucleolin in nucleus. Nucleolin was immunoprecipitated from NT2 subcellular fractions and co-precipitated RNAs were detected by qRT-PCR using primers for NLC1-C and β-actin (the negative control of cytoplasmic RNA) and U6 (the negative control of nuclear RNA). IP enrichment was determined as the amount of RNA associated to the Nucleolin IP relative to IgG control. Data are from one of three independent experiments and are represented as mean±S.D. with n=3. (j) Western blotting shows the amount of Nucleolin in input, IgG and immunoprecipitated Nucleolin containing mRNP complexes from both nucleus and cytoplasm fractions. Lamin A/C and β-tubulin were used as markers for nucleus and cytoplasm fractions, respectively (k). The expression of Nucleolin was down-regulated in MA patients. Quantitative real-time PCR analysis of the expression of Nucleolin in MA patients and normal control