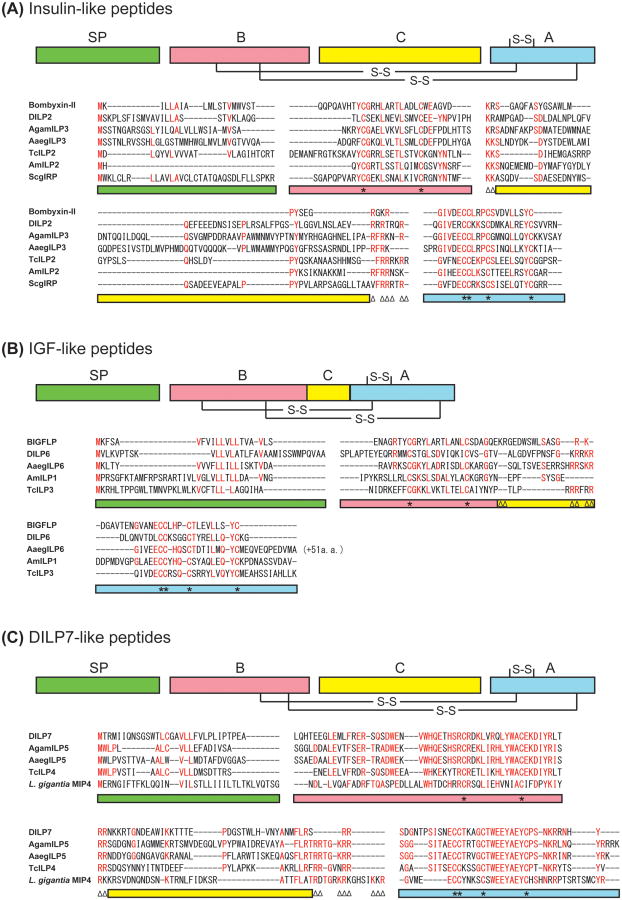
Figure 1.
Predicted insulin-like, IGF-like and DILP7-like peptides in insects. (A) Amino acid sequences of the representatives of predicted insulin-like peptides from Bombyx (bombyxin-II), Drosophila (DILP2), Anopheles (AgamILP3), Aedes (AaegILP3), Apis (AmILP2), Tribolium (TcILP2), and Schistocerca (ScgIRP) are aligned. (B) Amino acid sequences of the representatives of predicted IGF-like peptides from B. mori (BIGFLP), Drosophila (DILP6), Aedes (AaegILP6), Apis (AmILP1), and Tribolium (TcILP3) are aligned. (C) Amino acid sequences of the representatives of predicted highly conserved ILP group (DILP7-like peptides) from Drosophila (DILP7), Anopheles (AgamILP5), Aedes (AaegILP5), Tribolium (TcILP4), and Lottia (molluscan insulin-related peptide 4, MIP4) are aligned. Highly conserved amino acid residues are shown in red. Color bars indicate the predicted domains in the precursor peptides: green, signal peptide; red, B-chain; yellow, C-peptide; blue, A-chain. Asterisks on the color bars below the alignment denote Cys residues, and paired triangles denote potential cleavage sites (dibasic amino acids).