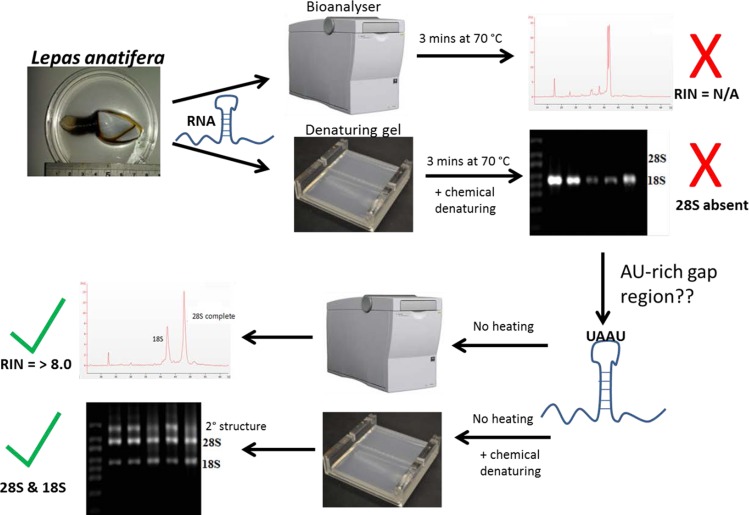
Figure 4. The steps involved in determining RNA quality in a species containing a ‘gap deletion’.
These include Arthropoda, most protostome animals and a scattering of other groups (-see Table 1). The schematic displays the problem with taking routine approaches in taxa with gap deletions because standard protocols specify heat-denaturation of the rRNA prior to Bioanalyser analysis. The latter measures migration and intensity of LSUs and RNA Integrity Numbers (RINs). Heat-denaturation prevents visualisation of the 28S peak in Bioanalyser electropherograms in the affected taxa, so that RNA appears ‘degraded’ and a RIN cannot be generated. The solution is to run non-heat-denatured rRNA during Bioanalyser analysis. Denaturing formaldehyde gel electrophoresis of rRNA does not appear to bring about the gap deletion and RNA subunits appear normally on these gels.