Figure 1.
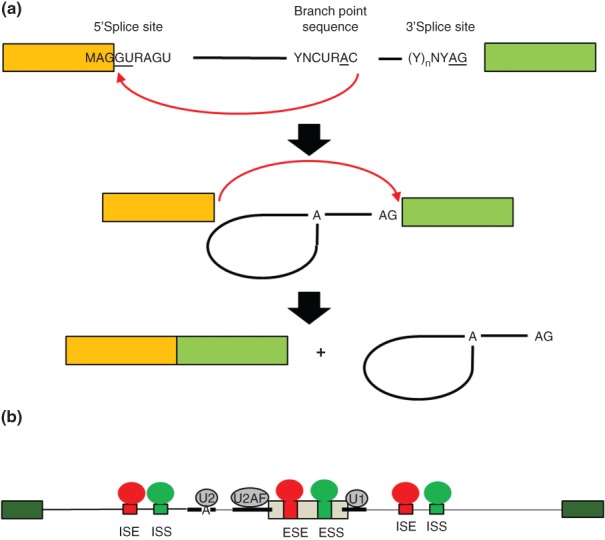
The biochemical steps of intron splicing and mechanisms of alternative splicing regulation. (a) Removal of an intron occurs in two catalytic steps that are directed by the core splice sites. In the first catalytic step of splicing, an adenosine residue in the branchpoint sequence (BPS) carries out nucleophilic attack at the 5′ end of the intron to form a branched 2′→5′ phosphodiester bond and lariat intermediate (middle). In the second catalytic step, the 3′ OH of the 5′ exon carries out nucleophilic attack at the 3′ splice site to ligate the exons in a 5′→3′ phosphodiester bond. The intron is released as a lariat product that is debranched and degraded. (b) Schematic of a model of combinatorial control of alternative splicing. An alternatively spliced cassette exon (gray) is flanked by two constitutively spliced exons (green). The positions of the core splice sites and BPS are indicated by thick lines. Intronic splicing enhancer (ISE) and exonic splicing enhancer (ESE) sequence elements are indicated in red together with splicing regulatory proteins (SRP) that bind them to promote exon splicing. Intronic splicing silencer (ISS) and exonic splicing silencer (ESS) are indicated in blue along with corresponding SRPs that promote exon skipping. The net combined activities of factors that promote splicing or skipping determine the level of exon splicing. These activities determine, at least in part, whether U1 binds to the 5′ splice site, U2AF to the PPT and 3′ splice site, and U2 to the BPS and subsequent steps of spliceosome assembly.
