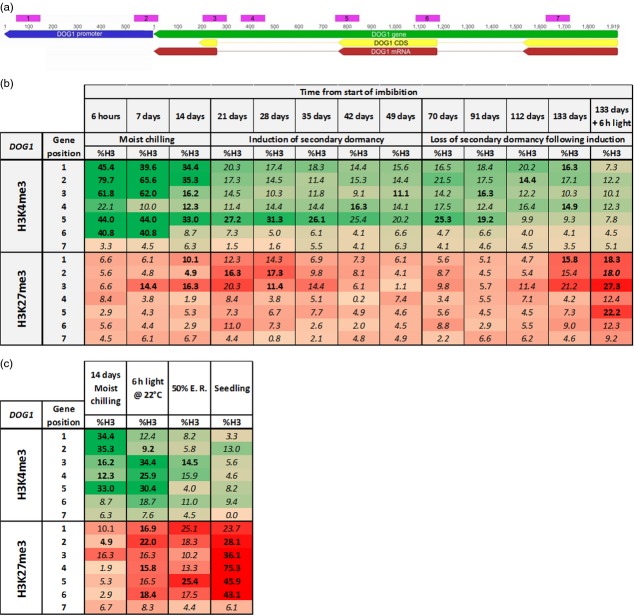
Figure 6.
Dynamic changes in H3K4me3 and H3K27me3 marks on DOG1 during dormancy cycling and germination.(a) Map of DOG1 gene structure showing the seven positions targeted by primers in qPCR of DNA from the ChIP assay (Pink regions). Primers targeted to regions in which the University of California Santa Cruz Genome Browser (http://epigenomics.mcdb.ucla.edu) indicated the presence of H3K4me3 and H3K27me3 marks in seedlings.(b) Heat map showing changes (%H3, defined below) in the activating histone modification H3K4me3 (Green heat map) and the repressive modification H3K27me3 (Red heat map) at seven positions on DOG1 during the decrease in primary dormancy in response to moist chilling, secondary dormancy induction and secondary dormancy loss as determined by chromatin immunoprecipitation followed by qPCR.(c) Heat map showing changes in the activating H3K4me3 (green heat map) and the repressive H3K27me3 (red heat map) modifications at seven positions on DOG1 following the decrease of primary dormancy through moist chilling and subsequent removal of the final layer of dormancy by exposure to 6 h light at 22°C to induce germination, the progression of germination to 50% endosperm rupture (50% E. R.) and in seedlings. Means of three biological replicates are shown. Values were normalised to the immunoprecipitates of unmodified H3 to account for variability in histone density. Numbers in bold are significantly higher (P < 0.05) than background noise, which was determined by ChIP with unspecific IgG. %H3 is the fraction of modified histone H3 (as determined with the antibody against the specific methylation) within total histone H3 (as determined with an IP using an antibody against all histone H3). Colour intensity is maximum for values of 30 or greater. Standard errors and P-values for all means are shown in Table S5.