Figure 1.
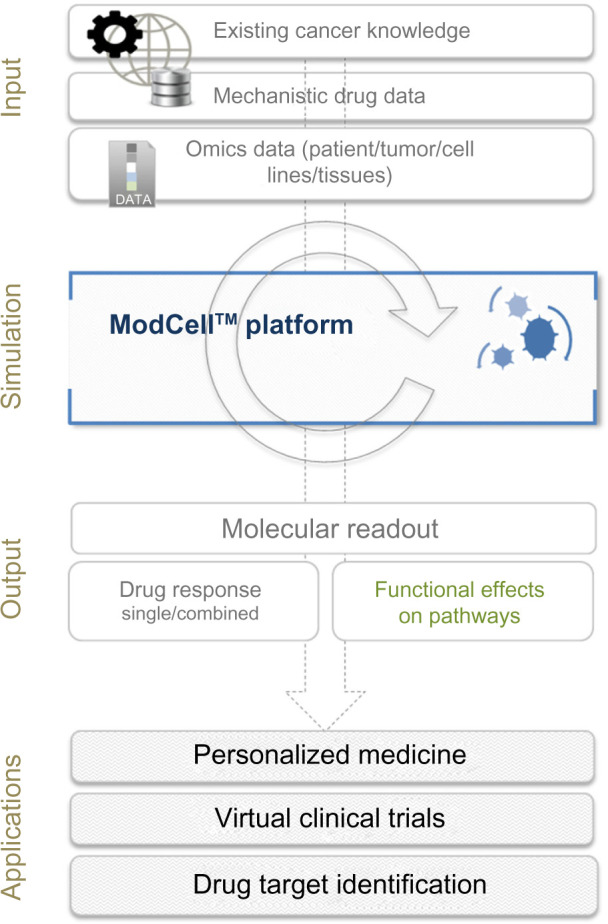
Overview of the ModCell™ predictive modeling approach in oncology. ModCell™ uses publicly available resources, representing the sum of knowledge on cancer, cell signaling, and drug action (eg, dissociation constants and molecular targets), to construct a large-scale mechanistic model of cellular signaling. A generic large-scale signaling network is established, which can be personalized with omics data (eg, transcriptome/exome/proteome) from individual patient tumors/cell lines/experimental tissues (public and/or private data resources). The effects of identified molecular alterations on pathway function and cross-talk can then be simulated using the mechanistic modeling approach implemented by ModCell™ and the underlying PyBioS modeling framework. Response to molecularly targeted drugs (single or in combination) can be predicted through establishment of a molecular readout (eg, MYC levels, phosphorylation status of TP53, cleavage of PARP1, and GTP loading status of RAC1 and CDC42) as a proxy for phenotypic effects (eg, cell proliferation, senescence, apoptosis induction and cell migration), allowing identification of the optimal treatment.
