Abstract
Introduction
Yellow fever continues to be a problem in sub-Saharan Africa with repeated epidemics occurring. The mosquito Aedes bromeliae is a major vector of yellow fever, but it cannot be readily differentiated from its non-vector zoophilic sister species Ae. lilii using morphological characters. Genetic differences have been reported between anthropophilic Ae. bromeliae and zoophilic Ae. lilii and between forest and domestic populations. However, due to the application of different molecular markers and non-overlapping populations employed in previous studies, interpretation of species delimitation is unclear.
Methodology/Principle Findings
DNA sequences were generated from specimens of Ae. simpsoni s.l. from the Republic of Benin, Tanzania and Uganda for two nuclear genes apolipophorin 2 (apoLp2) and cytochrome p450 (CYPJ92), the ribosomal internal transcribed spacer region (ITS) and the mitochondrial cytochrome c oxidase (COI) barcoding region. Nuclear genes apoLp2 and CYPJ92 were unable to differentiate between species Ae. bromeliae and Ae. lilii due to ancestral lineage sorting, while ITS sequence data provided clear topological separation on a phylogeny. The standard COI barcoding region was shown to be subject to species introgression and unable to clearly distinguish the two taxa. Here we present a reliable direct PCR-based method for differentiation of the vector species Ae. bromeliae from its isomorphic, sympatric and non-biomedically important sister taxon, Ae. lilii, based on the ITS region. Using molecular species verification, we describe novel immature habitats for Ae. lilii and report both sympatric and allopatric populations. Whereas only Ae. lilii is found in the Republic of Benin and only Ae. bromeliae in Tanzania, both species are sympatric in Uganda.
Conclusions/Significance
Our accurate identification method will allow informed distribution and detailed ecological studies that will facilitate assessment of arboviral disease risk and development of future targeted vector control.
Author Summary
In Africa, epidemic outbreaks of yellow fever continue despite the availability of an effective vaccine. Effective understanding of disease epidemiology and control requires the ability to reliably identify vectors of yellow fever. The mosquito Ae. bromeliae, a competent vector of yellow fever virus, cannot be reliably morphologically differentiated from its sister species Ae. lilii, which does not bite humans and so does not transmit yellow fever. DNA sequencing of four molecular markers allowed comparisons of how they perform at distinguishing these species. We found that the mitochondrial cytochrome c oxidase (COI) barcoding region and nuclear apolipophorin 2 (apoLp2) and cytochrome p450 (CYPJ92) were unable to reliably distinguish these species. Conversely, genetic variation at the internal transcribed spacer region (ITS) was able to confirm the vector Ae. bromeliae and non-vector Ae. lilii as distinct species. Based on ITS sequence differences, we developed a robust molecular method to identify the vector Ae. bromeliae from its sister species Ae. lilii. Consequently, we find that these species use the same larval habitats including banana, cocoyam and Dracena spp. in Uganda. Whereas only Ae. lilii appears to be present in Benin and only Ae. bromeliae in Tanzania, we confirm that both species occur in Uganda. Reliable species designation will promote more detailed studies of distribution, ecology and vector status essential for disease risk assessment and mosquito control.
Introduction
Correctly identifying the vector species involved in mosquito-borne disease transmission is fundamental to predicting disease outbreaks, ascertaining general risk to the human population and targeting control efforts. Despite this necessity, the reliable identification of mosquitoes is problematic in many cases, including the medically important Aedes simpsoni complex. This complex comprises three known species including Ae. simpsoni, Ae. lilii and Ae. bromeliae. Among these three species, Ae. bromeliae is an important vector of yellow fever virus (YFV) and potentially other arboviruses [1]. Yellow fever has increased in incidence as a result of urbanisation and changes to public health policy [2, 3]. The burden of yellow fever in Africa is estimated to be 130,000 cases a year, 85,000 of which result in deaths, despite the availability of a vaccine [4]. After mass immunisation campaigns at the beginning of the 20th Century, YFV was successfully reduced in targeted countries. However, YFV outbreaks are causing renewed attention. In West Africa, 13 out of 14 countries known to host YFV now report cases regularly and have experienced epidemics since 2000 [5, 6]. Further concern has arisen over identification of a novel YFV genotype implicated in recent outbreaks including the first reported outbreak in Kenya, East Africa (1992–1993) [7]. Related genotypes have also been reported in Sudan in 2003 and 2005 and more recently in northern Uganda in 2010 [8–10]. Although much attention has been given to understanding yellow fever disease epidemiology [11], relatively little is known about the mosquito vectors.
Ten species are currently known within the wider Simpsoni Group (Huang 2004). Within this group, Theobald [12–14] originally described three species belonging to the Simpsoni Complex (Ae. simpsoni, Ae. bromeliae and Ae. lilii). Despite these separate species designations, many entomologists referred to all simpsoni-like mosquitoes within this complex as the nominotypical species, Ae. simpsoni [15–21]. In Uganda, it was found that some populations of Ae.simpsoni s.l. were anthropophilic and attracted to human bait, whereas others were not attracted to man despite local abundance [20]. Furthermore, mosquitoes collected from zoophilic and anthropophilic populations showed different feeding preferences in the laboratory with a preference for rodents or humans, respectively [20]. These findings led to a re-examination of mosquito morphology and subsequently Huang [22, 23] provided a full description of the component members of the Simpsoni Complex, reviewed their ecology and drew attention to the incorrect use of former nomenclature. Aedes simpsoni, which has only been reported from South Africa and Swaziland, is not implicated in human disease transmission [22, 23]. The anthropophilic yellow fever vector Ae. bromeliae is widespread on the African continent [23]. In contrast, Ae. lilii, only previously reported from Sudan, Ethiopia and Uganda, has never been reported biting man and is thus not considered to be involved in disease transmission [22, 23]. Although we know these three species have different distributions, biting behaviour and vectorial abilities, these need to be fully characterised. Understanding ecology and epidemiology requires that species can be distinguished from one another. However, controversy over mosquito taxonomy means that the anthropophilic disease transmitting Ae. bromeliae cannot be reliably distinguished from its zoophilic sister species Ae. lilii [24, 25].
Large scale detailed studies of the Simpsoni Complex by Huang [22, 23, 26], reported that Ae. simpsoni s.s. can be distinguished from Ae. bromeliae and Ae. lilii in that it has simple claws on the mid tarsi, as opposed to toothed mid-tarsal claws present in the latter two taxa. Aedes simpsoni can also be easily distinguished from conspecifics by its distinct tarsomere scaling pattern [22, 25]. However, Jupp & Kemp [24] reported variation in tarsal claw morphology of Ae. simpsoni and Ae. bromeliae, and questioned the reliability of this diagnostic character. In this study, we focus on the more widespread taxa of the complex, Ae. bromeliae and Ae. lilii. These sister taxa can sometimes be reliably identified morphologically based on tarsomere banding patterns, but only when this character is exhibited at the extremes of its range as banding patterns overlap between the species [25]. Lutwama & Mukwaya [25] questioned the usefulness of tarsomere banding patterns as a diagnostic character for Ae. bromeliae and Ae. lilii. They observed variation in scale ornamentation on an almost continuous scale and found that progeny from the same mother could be identified both as Ae. bromeliae or Ae. lilii based on this morphological character.
The unreliability and practicality of using morphological based methods for routine field-based identification in the Simpsoni Complex has led to attempts to delimit species boundaries through molecular methods. In a study of the Simpsoni Complex, excluding the southerly distributed Ae. simpsoni, Mukwaya et al. [27] found that anthropophilic populations from Kenya and Uganda form a distinct genetic clade separate from non-sympatric and non-anthropophilic populations from Uganda and Nigeria based on the non-coding internal transcribed spacer (ITS) region of ribosomal DNA. In that study, species were designated according to both blood feeding preference (because Ae. lilii is zoophilic whereas Ae. bromeliae are human vectors) and on tarsomere banding patterns which were distinctive for some specimens [22, 23, 27]. Recently, Walter et al. [28] inferred the presence of Ae. bromeliae and Ae. lilii in sympatry in Rabai, Kenya based upon genetic differences between domestic/peri-domestic and forest populations. However, how these putative species relate to those characterised by Mukwaya et al. [27] remains unclear as they used different molecular markers; two nuclear genes, apolipophorin 2 and cytochrome p450 (CYPJ92).
A 658 bp region of the cytochrome c oxidase (COI) gene has been widely adopted as a DNA barcoding standard for species identification because it shows high utility in discriminating between closely related taxa as well as resolving phylogeographic groups within species [29–31]. However, sole use of mitochondrial DNA to delimit species has been questioned because of the potential for pseudogene development and introgression, which may limit the ability of mtDNA markers to resolve closely related species [32, 33]. Combined analysis of both mitochondrial and nuclear genes can improve phylogenetic resolution since these markers evolve at different rates and so target different levels of the phylogenetic tree [34]. Another barcoding candidate is the nuclear internal transcribed spacer (ITS) regions of the ribosomal gene cistron that comprises the 18S, 5.8S, and 28S genes, an external spacer region and two internal spacer regions ITS1 and ITS2 [35]. The ITS regions evolve at a rapid rate in the absence of functional constraint [36, 37]. Because they are tandemly repeated in the genome, the ITS spacer regions are also subject to concerted evolution whereby paralogues are homogenised by genetic exchange [36, 37]. Consequently, paralogues remain genetically similar within species while showing high levels of interspecific divergence [37]. Although not useful in all taxonomic groups, ITS can differentiate between sister species in a large number of cases and has been widely used to delimit closely related mosquitoes in Anopheles complexes [35, 38–42].
We seek to expand previous work on genetic differentiation of Ae. bromeliae and Ae. lilii in the Simpsoni Complex by determining how the putative species identified by Mukwaya et al. [27] and Walter et al. [28] relate to one another. We achieve this by sequencing the same mosquito samples at previously used molecular markers (apoLp2, CYPJ92 and ITS). In addition, we were able to evaluate the utility of various molecular markers, including the ITS and COI regions, in determining species bounds. We apply these findings to develop a molecular identification method based on variation in the ITS region to distinguish the disease vector Ae. bromeliae from its non-vector sister species Ae. lilii. Applying this method to mosquitoes from Benin, Uganda and Tanzania enabled us to generate reliable findings of their ecology and distribution.
Methods
Sample collection
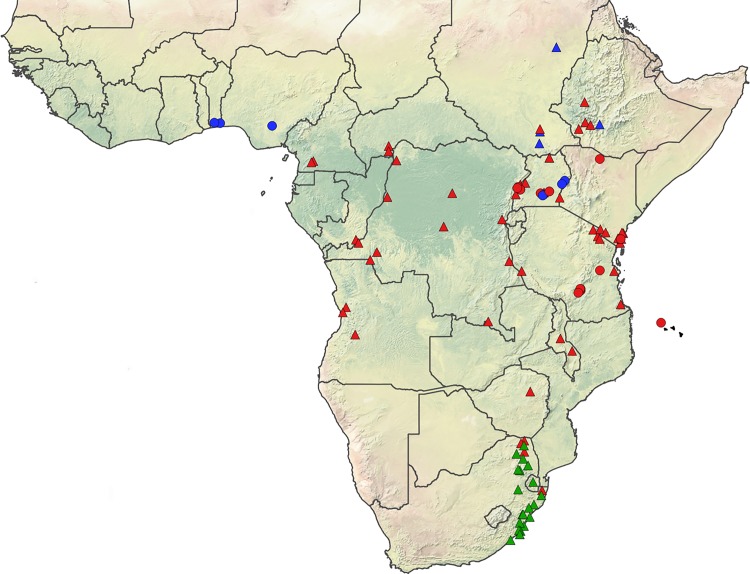
Mosquitoes of the Simpsoni Complex were collected as larvae from natural breeding sites in Tanzania (n = 36), Uganda (n = 50) and the Republic of Benin (n = 24) from locations detailed in Fig 1 and S1 Table from 2009 to 2014. Immature habitats sampled included the leaf axils of Musa spp. (banana), Colocasia spp. (cocoyam/taro), Dracaena spp. and in tree holes. To avoid biasing the dataset with siblings, each discrete habitat was treated as a separate collection and only one individual per collection was taken for genetic analyses. Where possible, immatures were reared though to adults; otherwise, larvae destined for DNA analysis were stored in 95% ethanol. Adults were desiccated with silica for optimal DNA preservation and either stored in BEEM capsules or pinned. Larvae were preserved in ethanol for later extraction of DNA. All adult mosquitoes were identified as belonging to the Simpsoni Complex using the morphological identification key in Huang [26], and a subsample of these, and outgroup taxa including Ae. aegypti and Ae. aegypti formosus, were morphologically verified by Dr. Yiau-Min Huang.
Fig 1. Map of sampling points for Ae. bromeliae (red circles) and Ae. lilii (blue circles) used in the current study including samples from Mukwaya et al. [27] and Le Goff et al. [43].
Also mapped are the previously described sampling points for Ae. bromeliae (red triangles), Ae. lilii (blue triangles) and Ae. simpsoni s.s (green triangles) based on morphological identification, taken from Huang [23]. The map was created in QGIS (QGIS Development Team) made with Natural Earth.
Experimental procedures
DNA was extracted from whole larvae or a single leg of an adult mosquito using the modified phenol-chloroform method in Surendran et al. [44]. Forty individuals of the Simpsoni Complex were amplified and sequenced for a region of the mitochondrial COI gene with universal primers LCO1490 (5’GGTCAACAAATCATAAAGATATTGG’3) and HCO2198 (5’TAAACTTCAGGGTGACCAAAAAATCA’3) [45] using a protocol recommended by the Consortium for the Barcode of Life (http://barcoding.si.edu/dnabarcoding.htm). Peridomestic Aedes aegypti collected from an artificial container in Tanzania was also sequenced at COI as an outgroup.
Internal transcribed spacer regions 1 and 2 were amplified from six individuals from Uganda and three individuals from Tanzania that were selected to represent the genetic diversity we observed at the COI gene. This was achieved with the 18SFHIN and CP16 primers (5’-GTAAGCTTCCTTTGTACACACCGCCCGT-3’ and 5’-GCGGGTACCATGCTTAAATTTAGGGGGTA-3’, respectively) [46], as used by Mukwaya et al. [27]. To generate PCR products, 1 unit of high fidelity MyFi DNA Polymerase (Bioline, UK), 2X MyFi Reaction Buffer, 0.8 μM forward and reverse primer and 1–10 ng of template DNA were used under the following conditions at 30% ramp speed; 95°C for 3 min followed by 30 cycles of 95°C for 30 sec, 58°C for 45 sec and 72°C for 45 sec with no final extension.
Thirteen Individuals, including eight of the same individuals sequenced at the ITS region, were also sequenced for regions of the nuclear genes apolipophorin 2 (apoLp2) and cytochrome p450 (CYPJ92) first described by Brown et al. [47] and used by Walter et al. [28] in the Simpsoni Complex. This included verified Aedes aegypti formosus collected from a tree hole in Tanzania, for use as an outgroup as was the case in Walter et al. [28]. Another marker, short-chain dehydrogenase-reductase (SDR) also used by Walter et al. [28] in the Simpsoni Complex was not used because it produced multiple nonspecific bands on amplification. PCR products were generated as in Walter et al. [28]. PCR products were purified with the GenElute PCR clean up kit (Sigma-Aldrich, UK) and Sanger sequenced in forward and reverse directions using the amplification primers. Sequences were generated with BigDye Terminator v3.1 cycle sequencing kit (Applied BioSystems, UK) on an Applied BioSystems 3730 automated sequencer.
ITS products were cloned using the P-GEM cloning kit (Promega, UK) as per instructions. Transformants were blue/white screened and colonies with inserts stored in TE buffer for PCR amplification. Universal M13 primers (5’-TGTAAAACGACGGCCAGT-3’ and 5’-CAGGAAACAGCTATGAC-3’) [48] were used to amplify cloned ITS products in the following 13μl reaction; 1.25 units of BIOTAQ DNA Polymerase (BioLine, UK), 1 X NH4 Reaction Buffer, 2mM MgCl2 solution, 0.8mM dNTP and 0.5 μM forward and reverse primer. Thermocycler conditions were 95°C for 2 min followed by 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec for 35 cycles and a final extension of 72°C for 10 min. A minimum of two and a maximum of four clones were forward sequenced for each individual.
Cloned ITS sequences were aligned with Mukwaya et al.’s [27] from several locations in Africa (S1 Table) and Le Goff et al.’s [43] sequence data from two Ae. bromeliae originating from the Indian Ocean island of Mayotte available from GenBank (KF135509-10) using the program Geneious v5.4.7 [49]. Primers were designed based on this alignment to discriminate between species. Several putative species-specific primers were trialled under a wide range of PCR conditions. However, the only primers that worked effectively generated species-specific PCR products that differed by ~30 b.p. Due to this small size difference we recommend that the primer pairs specific for each species are run in separate PCR reactions. Non-specific banding was a feature of all primers tested which we suspect may result from variable indel length among ITS copies within an individual. Although PCR protocols were optimised to reduce non-specific banding, some extra banding can be visible but does not obscure amplification/non-amplification of the species-specific PCR product.
The developed primers are nested within the ITS1-2 region; therefore PCR products encompassing this region were first generated with the 18SFHIN and CP16 primers as described above. These PCR products were purified with the GenElute PCR clean up kit (Sigma, UK) and 0.5 μl was used as template in a 25 μl reaction with 0.6 units of Go Taq Hot Start polymerase (Promega, UK), 1 X NH4 reaction buffer, 1 mM MgCl2 solution, 0.8 mM dNTP and 0.5 μM forward and reverse primer. Primers developed for amplification of 591 bp in Ae. bromeliae only were BRO-F (5’-CCTGGCCAGTGGCCA-3’) and BRO-R (5’-GTGCACACCACTGA-3’). Amplification was achieved with a touchdown PCR protocol; initialisation step of 95°C for 3 minutes followed by 95°C for 30 seconds, 82°C for 45 seconds and 72°C for 1 minute for 5 cycles, followed by 30 cycles of 95°C for 30 seconds, 64°C for 45 seconds and 72°C for 1 minute and then a final extension of 72°C for 7 minutes. Primers developed for amplification of a 620 bp region in Ae. lilii only were LIL-F (5’CTGATGCACTGGCCTCAAAG’3) and LIL-R (5’TCAACCGCCGTGCGTG’3). The thermocycling conditions were 95°C for 3 minutes followed by 95°C for 30 seconds, 78°C for 45 seconds, 72°C for 1 minute for 10 cycles followed by 95°C for 30 seconds, 70°C for 45 seconds, 72°C for 1 minute for 20 cycles and a final extension step at 72°C for 7 minutes. Amplified products were run on a 1.2% agarose electrophoresis gel to determine the presence or absence of DNA bands of the expected size. Positive and negative species controls were used in all PCR reactions. Sequences are available on GenBank (KT998333- KT998452).
Data analysis
Sequence alignment was achieved with Geneious v5.4.7 [49]. For the apoLp2 and CYPJ92 datasets, files were prepared using seqPHASE [50] and PHASE v2.1 was then used to infer haplotypes [51]. The haplotypes of Ae. aegypti formosus outgroups were determined through alignment with the relevant datasets for Ae. aegypti in Brown et al. [47] and using the program PHASE. Sequences for two Ae. aegypti from Brown et al. [47] were used as outgroups in conjunction with data generated during the present study. The COI dataset was aligned with two COI sequences of Ae. bromeliae from the Indian Ocean island of Mayotte available from GenBank (KF135496-97) [43].
Neighbour joining (NJ) trees were constructed in MEGA 6 [52]as in Mukwaya et al.[27] and Walter et al. [28] using the best available substitution model as chosen by JModelTest [53, 54]. The Tamura-Nei model with uniform rates among sites was used to construct NJ trees for COI. The Kimura 2 parameter model (K80) with uniform rates among sites was used for ITS sequences and K80 with 0.8 gamma-distributed sites was used to construct trees for apoLp2 and CYPJ92. For all genetic markers, missing data including indels were excluded from analysis. Topological support was determined through 1000 bootstrap replications. The ITS sequence tree was constructed without an outgroup because the high level of divergence between the outgroup and the ingroups presents a challenge for sequence alignment. The two ITS sequences of Ae. bromeliae from Mayotte [43] were not included in the NJ tree because they did not overlap with the sequences generated here.
A hierarchical AMOVA was performed on the ITS sequence data generated in this study together with Mukwaya et al.’s sequences [27] (n = 69) in Arlequin v3.5 [55]. A hierarchical AMOVA was also performed on the apoLp2 (n = 13) and CYPJ92 (n = 13) sequences generated in this study.
Results
Phylogenetic analysis
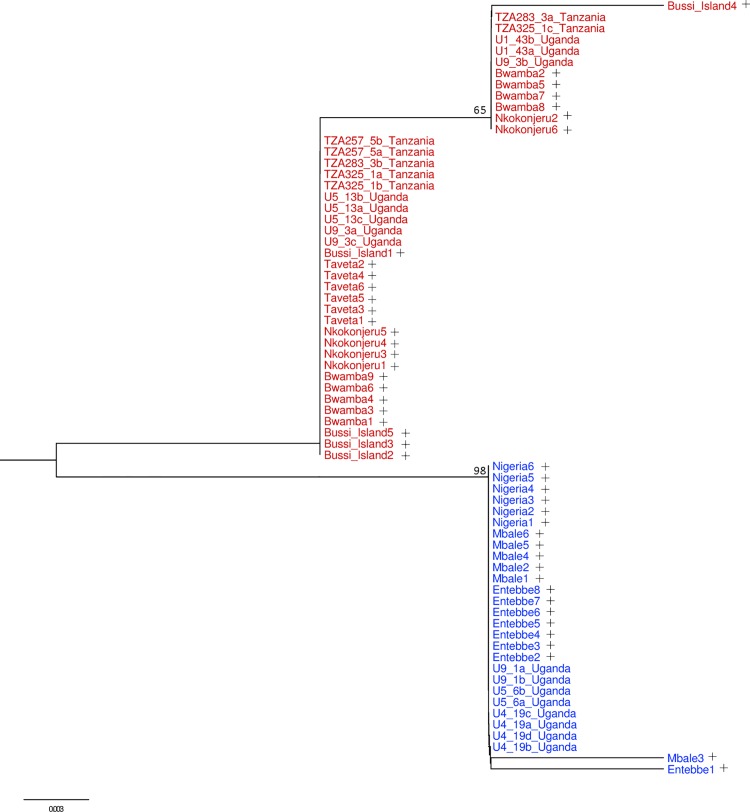
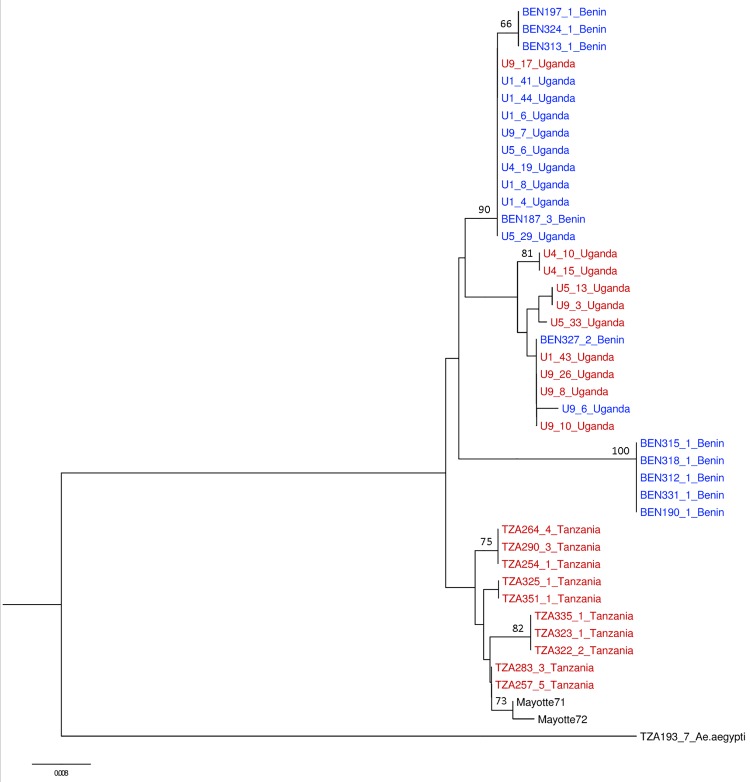
Four fixed point substitutions and eight indels of varying length were observed between species in the first 300 bp of the ITS sequence alignment. The neighbour joining tree of ITS sequences revealed two major clades with a bootstrap support of 98% (Fig 2). As reported previously, the 46 sequences from Mukwaya et al. [27] clustered into one or other of the two clades depending on their inferred host feeding preference and which they accordingly designated as Ae. bromeliae (anthropophilic) and Ae. lilii (non-anthropophilic). Sequences of all nine Simpsoni Complex individuals from this study also belonged to one or other of these clades, and we therefore identified them as Ae. bromeliae or Ae. lilii with reference to Mukwaya et al.’s [27] ITS-based species designation.
Fig 2. Neighbour joining tree of ITS sequence data with labels coloured according to species designation based on ITS as in Mukwaya et al. [27].
Red labels represent Ae. bromeliae while blue labels are Ae. lilii. Sequences from Mukwaya et al. [27] (+) are annotated. Bootstrap support values above 55% are shown.
Mosquitoes collected from Kanyawara, Bundibugyo (Bwamba region) and Najjembe in Uganda represented both ITS clades. In comparison, individuals collected from Tanzania strictly aggregated into the ITS lineage of Ae. bromeliae. AMOVA revealed that almost all sequence differences can be explained by variation between species (92.65%), while there is little variation between populations within groups and within populations (0.25% and 7.19%, respectively).
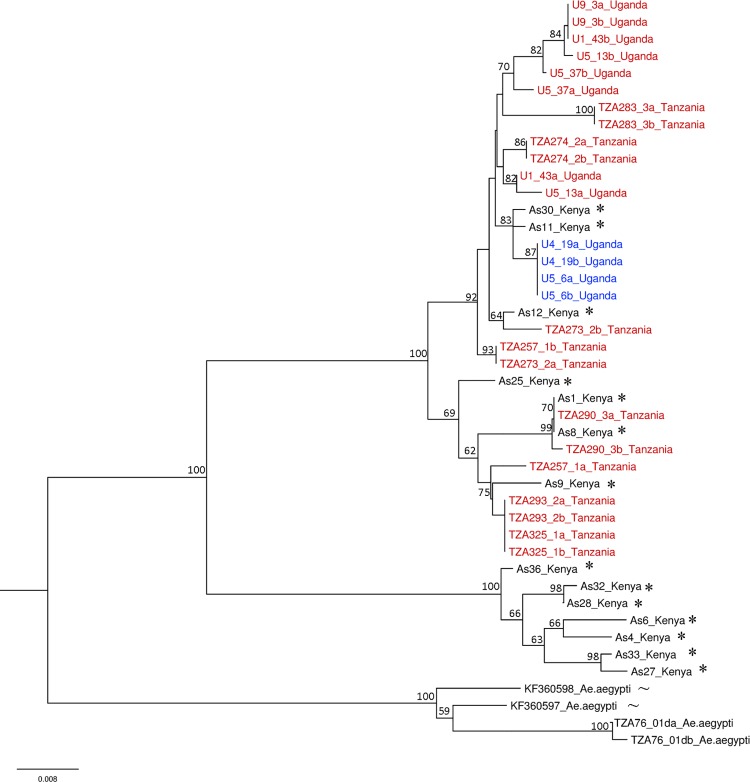
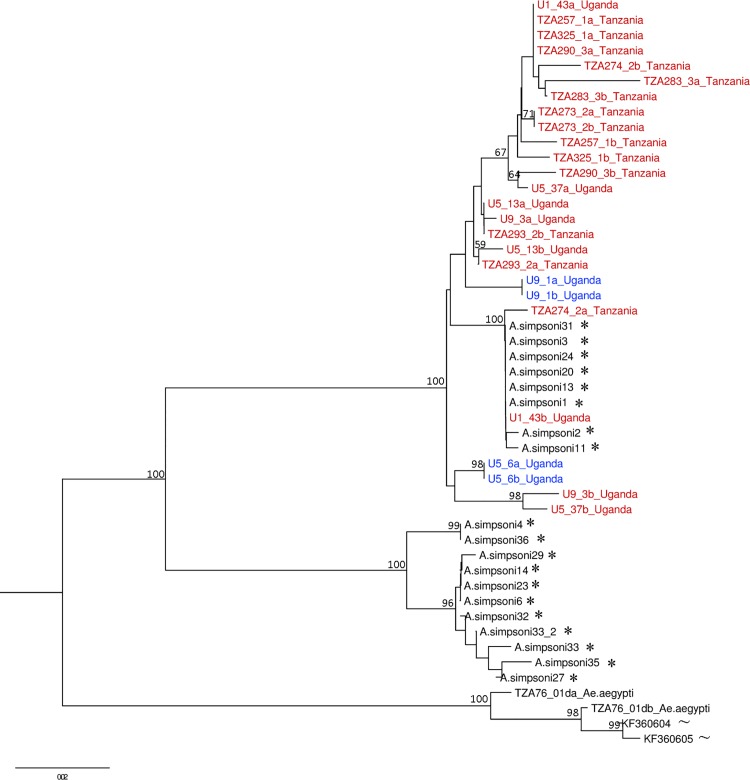
Sequencing of individuals at both ITS and nuclear genes apoLp2 and CYPJ92 allowed us to compare the ITS-based species designation of Mukwaya et al. [27] with that of Walter et al. [28]. Two well-supported lineages were observed in NJ trees for the nuclear genes apoLp2 and CYPJ92 with 100% bootstrap support (Figs 3 and 4). Individuals confirmed as Ae. bromeliae or Ae. lilii using the ITS region with reference to Mukwaya et al. [27], all fall into a single clade for each of the nuclear genes, apoLp2 and CYPJ92. For both gene trees, the other lineage comprises only the sequences from the forest species in Walter et al. [28] that they referred to as Ae. lilii. A hierarchical AMOVA revealed that nuclear genes apoLp2 and CYPJ92 could not effectively distinguish between Ae. bromeliae and Ae. lilii with only 0.86% and 18.94% of genetic variance between species groups, respectively. Conversely, there was relatively high genetic variation among populations within species groups (37.04%, 10.28%, respectively) and within populations (62.10%, 70.79%, respectively).
Fig 3. Neighbour joining tree of apoLp2 sequence data with labels coloured according to ITS-based species designation.
Red labels represent Ae. bromeliae while blue labels are Ae. lilii. Sequences from Walter et al. [28] (*) and Ae. aegypti outgroups (~) from Brown et al. [47] are annotated. Bootstrap support values for branches above 55% are shown.
Fig 4. Neighbour joining tree of CYPJ92 sequence data with labels coloured according to ITS-based species designation.
Red labels represent Ae. bromeliae while blue labels are Ae. lilii. Sequences from Walter et al. [28] (*) and Ae. aegypti outgroups (~) from Brown et al. [47] are annotated. Bootstrap support values for branches above 55% are shown.
There are two phylogenetic clusters with high bootstrap support (90%, 90%) and two with weak bootstrap support (56% and 43%) in the NJ tree for mitochondrial COI with individuals tending to cluster according to geographic origin (Fig 5). The sequences from Mayotte cluster with Ae. bromeliae from Tanzania. Individuals designated as Ae. bromeliae or Ae. lilii according to ITS sequence variation tend to group according to species within the same clades, but there are three exceptions. One individual from Uganda, exhibiting the ITS sequence of Ae. bromeliae clusters within a grouping of Ae. lilii while two individuals from Uganda and Benin, identified as Ae. lilii, cluster within a grouping that is otherwise comprised of Ae. bromeliae. A hierarchical AMOVA showed there is more genetic variation within (42.83%) and between populations (55.18%) than between species (1.99%) at the COI gene.
Fig 5. Neighbour joining tree of COI sequence data with labels coloured according to ITS-based species designation.
Red labels represent Ae. bromeliae while blue labels are Ae. lilii. Sequences from Le Goff et al. [43] (^) are annotated. Bootstrap support values for branches above 55% are shown.
Species identification and geographical distribution
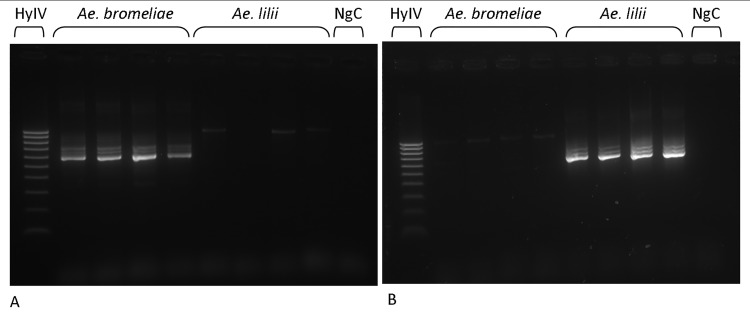
Based on fixed differences between species in the ITS sequences, species specific primers were designed to amplify PCR products in Ae. bromeliae or Ae. lilii. Primers designed to amplify in Ae. bromeliae did not amplify a PCR product from Ae. lilii and vice versa (Fig 6). Application of this method to 110 specimens of Ae. simpsoni s.l. positively identified all individuals as either Ae. bromeliae or Ae. lilii, as detailed in Table 1. Both species use the same breeding habitats in domestic and peridomestic habitats including the leaf axils of Musa spp., Colocasia spp. and Dracena spp. Despite focussed collection attempts, neither species was found utilising tree holes for immature development. Both species occur in sympatry in Uganda, while only Ae. lilii was collected from the Republic of Benin and Ae. bromeliae was the only species detected in Tanzania (Fig 1).
Fig 6. Electrophoresis gel for primer sets (A) BRO-F and BRO-R and (B) LIL-F and LIL-R.
PCR products were run with Hyperladder IV (HyIV) and a negative control (NgC).
Table 1. The number of individuals (n = 110) identified from sampled locations using our PCR mediated identification method.
| Country | Village/town | Ae. bromeliae | Ae. lilli | |
|---|---|---|---|---|
| Tanzania | Mlimba | 5 | 0 | |
| Udagaji village | 19 | 0 | ||
| Chita | 1 | 0 | ||
| Mahenge | 10 | 0 | ||
| Morningside | 1 | 0 | ||
| Total | 36 | 0 | ||
| Benin | Pobe | 0 | 14 | |
| Niaouili Village | 0 | 10 | ||
| Total | 0 | 24 | ||
| Uganda | Kanyawara | 1 | 14 | |
| Bundibugyo | 14 | 6 | ||
| Kapchorwa | 0 | 4 | ||
| Najjembe | 5 | 6 | ||
| Total | 20 | 30 | ||
Discussion
Here we used multiple markers to confirm there are two closely related species of the Simpsoni Complex that occur both allopatrically and sympatrically across the sampled range in sub-Saharan Africa. These species correspond to Ae. bromeliae and Ae. lilii of the Simpsoni Complex as characterised by Mukwaya et al. [22, 27]. We infer that a third forest taxon originally reported as Ae. lilii in Walter et al. [28] relates to another species, possibly a another member of the Simpsoni Group. The development of a molecular species identification method for the YFV vector Ae. bromeliae and non-vector Ae. lilii using variation at the ITS region allowed us to make for the first time reliable inferences about mosquito ecology and distribution. Such information is vital for a complete understanding of disease transmission by these species.
All the Ae. simpsoni s.l. mosquitoes tested fell into one or other of two genetically divergent ITS lineages. Based on sequence similarity these correspond directly to the two lineages found by Mukwaya et al. [27] and we follow their previous designation as Ae. bromeliae and Ae. lilii. We consider their species designation to be reliable as it was based on both morphology and feeding preference. Firstly, even though some individuals could not be identified due to overlap in morphological characters, Mukwaya et al. [27] were able to identify some individual specimens from the defining characters of leg tarsomere banding pattern and claw morphology described by Huang [22, 23]. Secondly, Mukwaya et al. [27] identified species based upon host feeding behaviour; human landing catches were used to distinguish anthropophilic Ae. bromeliae from non-anthropophilic Ae. lilii which were collected as larvae where human biting members of the Simpsoni Complex were absent. We confirm the existence of Ae. bromeliae and Ae. lilii as two distinct species since, using ITS sequence variation, they remain genetically distinct in sympatry at several locations in Uganda.
Our study enabled us to link the work of Mukwaya et al. [27] with that of Walter et al. [28]. Whereas Muwakya et al. [27] focused on feeding behaviour, Walter et al. [28] found divergence between forest and peridomestic populations in coastal Kenya at three nuclear genes. Our phylogenetic analysis reveals that the genetic variation in the ITS region does not correspond to the two distinct lineages found in Walter et al. [28]. Both Ae. bromeliae and Ae. lilii fall into a single phylogenetic clade at nuclear genes apoLp2 (apolipophorin 2) and CYPJ92 (cytochrome p450). This is consistent with these more slowly evolving nuclear genes being unable to resolve recently diverged species due to incomplete lineage sorting [56]. The forest lineage from Kenya detected by nuclear genes apoLp2 and CYPJ92 is clearly distinct and has no shared ancestral polymorphism with either Ae. bromeliae or Ae. lilii indicating a more distant relationship. Morphological identification of mosquito species is notoriously difficult and there are many species within the wider Simpsoni Group which share morphological characteristics [26]. We therefore suggest that the forest species reported by Walter et al. [28] represents a member of the wider Simpsoni Group, rather than the Simpsoni Complex. Possible candidates for this species that are morphologically similar and known from Kenya include Ae. gandaensis, Ae. woodi, Ae. subargenteus and Ae. sampi [26].
The mitochondrial COI species barcode marker was also unable to distinguish these taxa with most genetic variation occurring between geographic populations rather than between species. Whilst this inability of COI to distinguish Ae. bromeliae and Ae. lilii could be due to incomplete lineage sorting, the phylogeny suggests it is most likely due to mtDNA introgression; there are three cases in which Ae. bromeliae clusters with Ae. lilii or vice versa, two of which occur in sympatry in Uganda. Mitochondrial introgression is fairly common between closely related mosquito taxa [33]. Given this, and that Aedine mosquitoes may be particularly prone to NUMT’s due to their large genome size [57] it would seem wise to not rely on mitochondrial markers to distinguish species in this genus without prior confirmation from an additional marker. We found that the ITS region was the only marker able to reliably separate species of the Simpsoni Complex and therefore used it to develop a PCR mediated species identification method. This method is widely applicable across the range of Ae. bromeliae and Ae. lilii except in southern Africa. For use in this region, the method should be modified to accommodate the presence of Ae. simpsoni s.s. Our PCR mediated species diagnostic method removes the difficulties imposed by morphological identification of field specimens and therefore provides a valuable asset to medical entomologists studying arbovirus transmission.
Our identification tool in conjunction with larval sampling from natural habitats has provided further information on the ecology of the Simpsoni Complex. Contrary to the wide range of plant species utilised by Ae. bromeliae as breeding habitats, the immatures of Ae. lilii have only been reported to date from the axils of Sansevieria spp., suggesting a narrower range of immature habitats [22, 23]. However, we found that Ae. lilii, like Ae. bromeliae, utilised the plant axils of Musa spp., Colocasia spp. and Dracena spp., suggesting that larval breeding sites are not as restricted as previously reported [22, 23]. Walter et al. [28] hypothesised that selective pressure for use of domestic larval habitats may have driven speciation in the Simpsoni Complex. This is not supported by our findings that showed no obvious differences in larval habitat between species, although our characterisation of larval habitats was not exhaustive. As host choice appears to be an ecological difference between these species, it is possible that divergent selection for anthropophily, and the reliable blood source it provides, could have driven species divergence [28, 58].
We have shown here that Ae. bromeliae and Ae. lilii are common in Uganda where they can be found in sympatry in peridomestic habitats. Ae. lilii was the only member of the Simpsoni Complex collected from the Republic of Benin where it is described for the first time, while only Ae. bromeliae was collected in Tanzania. In addition, Mukwaya et al.’s [27] sequence data shows that mosquitoes from Nigeria are Ae. lilii whereas those from Kenya are Ae. bromeliae. Molecular evidence therefore agrees with the earlier morphological data of Huang [22, 23] that Ae. bromeliae is prevalent in East Africa (Tanzania, Kenya, Uganda, Mayotte; Fig 1). This morphological data also indicates a wider distribution across sub-Saharan Africa for Ae. bromeliae (Fig 1) [22, 23], but it would be wise to confirm this using molecular identification. However, given that Ae. bromeliae tends to be readily collected when it is present, the molecular data suggest that only Ae. lilii is present in West Africa (Fig 1). This difference would be consistent with the early reports that West African populations are predominantly non-anthropophilic [59]. Based on our preliminary distribution data, Ae. bromeliae may be an important vector of yellow fever in East Africa where YFV has been isolated from the Simpsoni Complex previously and implemented in disease epidemics including the Ethiopian outbreak of 1960–61 [59–61]. In comparison, other mosquito vectors including Ae. aegypti, Ae. luteocephalus, Ae. furcifer and Ae. taylori may be more important for YFV disease transmission in West Africa where yellow fever has been isolated from these vectors and implicated in outbreaks [59, 62–65].
Emerging/re-emerging arboviruses such as Chikungunya and Zika are causing great concern since they are increasingly responsible for catastrophic epidemics worldwide [66–72]. Members of the Simpsoni Group transmit a range of arboviruses including yellow fever, Babanki and Ngari viruses [1]. A limited number of arboviral studies have focused on disease transmission in this species group, progress of which is hampered by incomplete taxonomic understanding. There is therefore a great need to resolve the molecular systematics of the wider Simpsoni Group that should in turn be used to develop methods of species identification. In addition to studies of arboviral risk, assessment of mosquito species ranges is required in order to relate these to differences in the distribution of arboviruses and/or arboviral genotypes such as that observed for yellow fever [59]. Improved identification methods would also facilitate studies on feeding behaviour and genetic introgression which are important for understanding the risks of zoonotic disease emergence. For example, the introgression we observed between Ae. lilii and Ae. bromeliae could increase the propensity of Ae. lilii to feed on humans, resulting in the increased transfer of zoonotic disease into humans. A precedent for this can be seen in Culex pipiens in North America where mixing of the molestus and pipiens genetic forms (predominantly human and bird feeding, respectively) increases transmission of West Nile virus to humans. A wide range of inferences on ecology and epidemiology can now be made with our molecular identification tool that should be used to assess disease risk and provide basic information for vector population control.
Supporting Information
(XLSX)
Acknowledgments
The authors are grateful to field teams at the Uganda Virus Research Institute and International Institute for Tropical Agriculture for their research assistance.
Data Availability
All sequence files are available from the GenBank database (accession number(s) KT998333- KT998452).
Funding Statement
This work was supported by an Africa Award from the Royal Society and Leverhulme Trust (AA110092) and a PhD studentship to KLB funded by the Natural Environment Research Council [NE/H525170/1,NE/1528134/1, NE/J500057/1]. This manuscript was prepared whilst YML held a National Research Council (NRC) Research Associateship Award at the Walter Reed Army Institute of Research. The material to be published reflects the views of the authors and should not be construed to represent those of the Department of the Army or the Department of Defense. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mutebi JP, Crabtree MB, Kading RC, Powers AM, Lutwama JJ, Miller BR. Mosquitoes of Western Uganda. J Med Entomol. 2012;49(6):1289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gubler DJ. Resurgent vector-borne diseases as a global health problem. Emerg Infect Dis. 1998. Jul-Sep;4(3):442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tomori O. Yellow fever in Africa: Public health impact and prospects for control in the 21st century. Biomedica. 2002;22(2):178–210. [PubMed] [Google Scholar]
- 4. Garske T, Van Kerkhove MD, Yactayo S, Ronveaux O, Lewis RF, Staples JE, et al. Yellow fever in Africa: Estimating the burden of disease and impact of mass vaccination from outbreak and serological data. PLoS Med. 2014;11(5):e1001638 10.1371/journal.pmed.1001638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. Global alert and response: The West African situation. http://www.who.int/csr/disease/yellowfev/westafrica/en/ 2015.
- 6. Roberts L. Resurgence of yellow fever in Africa prompts a counterattack. Science. 2007;316(5828):1109–. [DOI] [PubMed] [Google Scholar]
- 7. Ellis BR, Barrett ADT. The enigma of yellow fever in East Africa. Rev Med Virol. 2008;18(5):331–46. 10.1002/rmv.584 [DOI] [PubMed] [Google Scholar]
- 8. Onyango CO, Grobbelaar AA, Gibson GVF, Sang RC, Sow A, Swanepoel R, et al. Yellow fever outbreak, Southern Sudan, 2003. Emerg Infect Dis. 2004;10(9):1668–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gould LH, Osman MS, Farnon EC, Griffith KS, Godsey MS, Karch S, et al. An outbreak of yellow fever with concurrent chikungunya virus transmission in South Kordofan, Sudan, 2005. Trans R Soc Trop Med Hyg. 2008;102(12):1247–54. 10.1016/j.trstmh.2008.04.014 [DOI] [PubMed] [Google Scholar]
- 10. Wamala JF, Malimbo M, Okot CL, Atai-Omoruto AD, Tenywa E, Miller JR, et al. Epidemiological and laboratory characterization of a yellow fever outbreak in northern Uganda, October 2010–January 2011. Int J Infect Dis. 2012;16(7):e536–e42. 10.1016/j.ijid.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 11. Barrett ADT, Higgs S. Yellow fever: A disease that has yet to be conquered. Annu Rev Entomol. 2006;52(1):209–29. [DOI] [PubMed] [Google Scholar]
- 12. Theobald FV. A new Stegomyia from transvaal. Entomologist. 1905. (38):101–4. [Google Scholar]
- 13. Theobald FV. A monograph of the Culicidae or mosquitoes British Museum of Natural History, London; 1910. [Google Scholar]
- 14.Theobald FV. Uganda Culicidae including thirteen new species. In Novae Culicidae, Part 1. Wye, Kent; 1915.
- 15. Haddow AJ. A note on the occurrence of Aedes (Stegomyia) simpsoni Theobald in the canopy of rain-forest in Bwamba County, Uganda. Ann Trop Med Parasitol. 1950. October;44(3):238–41. [DOI] [PubMed] [Google Scholar]
- 16. Haddow AJ. The natural history of yellow fever in Africa. Proceedings of the Royal Society of Edinburgh Section B: Biological Sciences. 1969;70(03):191–227. [Google Scholar]
- 17. Gillett JD. The habits of the mosquito Aedes (Stegomyia) simpsoni (Theobald) in relation to the epidemiology of yellow fever in Uganda. Ann Trop Med Parasitol. 1951;45(2):110–21. [DOI] [PubMed] [Google Scholar]
- 18. Gillett JD. Further studies on the biting behaviour of Aedes (Stegomyia) simpsoni Theobald in Uganda. Ann Trop Med Parasitol. 1955;49(2). [DOI] [PubMed] [Google Scholar]
- 19.Mukwaya LG. Host preference in Aedes (Stegomyia) species mosquitoes with special preference to the anthropophilic forms of Ae (Stegomyia) simpsoni Theo. (Diptera: Culicidae) in Uganda: PhD thesis. University of East Africa; 1971.
- 20. Mukwaya LG. Genetic control of feeding preferences in the mosquitoes Aedes (Stegomyia) simpsoni and aegypti . Physiol Entomol. 1977;2(2):133–45. [Google Scholar]
- 21. Mahaffy AF, Smithburn KC, Jacobs HR, Gillett JD. Yellow fever in Western Uganda. Trans R Soc Trop Med Hyg. 1942;36(1):9–20. [Google Scholar]
- 22. Huang Y-M. Aedes (Stegomyia) simpsoni complex in the Ethiopian region with lectotype designation for simpsoni (Theobald) (Diptera: Culicidae). Mosq Syst. 1979;11(3):221–34. [Google Scholar]
- 23. Huang YM. Aedes (Stegomyia) bromeliae (Diptera: Culicidae), the yellow fever virus vector in East Africa. J Med Entomol. 1986;23(2):196–200. [DOI] [PubMed] [Google Scholar]
- 24. Jupp PG, Kemp A. Variation in tarsal claw morphology and the identification of Aedes (Stegomyia) demeilloni/segermanae and Aedes (Stegomyia) simpsoni/bromeliae (Diptera: Culicidae) in South Africa. J Am Mosq Control Assoc. 1999;15(1):86–8. [PubMed] [Google Scholar]
- 25. Lutwama JJM, Louis Godfrey Variation in morphological characters of adults of the Aedes (Stegomyia) simpsoni complex from Uganda, Kenya and South Africa (Dipteria: Culicidae). Mosq Syst. 1994;26(3):145–57. [Google Scholar]
- 26. Huang Y-M. The subgenus Stegomyia of Aedes in the Afrotropical region with keys to the species (Diptera: Culicidae). Zootaxa. 2004;700:1–120. [Google Scholar]
- 27. Mukwaya LG, Kayondo J. K., Crabtree M. B., Savage H. M., Biggerstaff B. J., Miller B. R. Genetic differentiation in the yellow fever virus vector, Aedes simpsoni complex, in Africa: sequence variation in the ribosomal DNA internal transcribed spacers of anthropophilic and non-anthropophilic populations. Insect Mol Biol. 2000;9(1):85–91. [DOI] [PubMed] [Google Scholar]
- 28. Walter KS, Brown J. E., Powell J. R. Microhabitat partitioning of Aedes simpsoni (Diptera: Culicidae). J Med Entomol. 2014;51(3):596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences. 2003;270(1512):313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hebert PDN, Ratnasingham S, de Waard JR. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London B: Biological Sciences. 2003;270(Suppl 1):S96–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Linton Y-M, Pecor JE, Porter CH, Mitchell LB, Garzón-Moreno A, Foley DH, et al. Mosquitoes of eastern Amazonian Ecuador: Biodiversity, bionomics and barcodes. Mem Inst Oswaldo Cruz. 2013;108(Suppl 1):100–9. 10.1590/0074-0276130440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Funk DJ, Omland KE. Species-level paraphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics. 2003;34(1):397–423. [Google Scholar]
- 33. Walton C, Sharpe RG, Pritchard SJ, Thelwell NJ, Butlin RK. Molecular identification of mosquito species. Biol J Linn Soc. 1999;68(1–2):241–56. [Google Scholar]
- 34. Huelsenbeck JP, Bull JJ, Cunningham CW. Combining data in phylogenetic analysis. Trends Ecol Evol. 1996;11(4):152–8. [DOI] [PubMed] [Google Scholar]
- 35. Collins FH, Paskewitz SM. A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol Biol. 1996;5(1):1–9. [DOI] [PubMed] [Google Scholar]
- 36. Arnheim N, Krystal M, Schmickel R, Wilson G, Ryder O, Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proceedings of the National Academy of Sciences. 1980;77(12):7323–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liao D. Concerted evolution: Molecular mechanism and biological implications. Am J Hum Genet. 1999;64(1):24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yao H, Song J, Liu C, Luo K, Han J, Li Y, et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE. 2010;5(10):e13102 10.1371/journal.pone.0013102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A. 2005;102(23):8369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walton C, Somboon P, O’Loughlin SM, Zhang S, Harbach RE, Linton YM, et al. Genetic diversity and molecular identification of mosquito species in the Anopheles maculatus group using the ITS2 region of rDNA. Infect, Genet Evol. 2007;7(1):93–102. [DOI] [PubMed] [Google Scholar]
- 41. Prakash A, Walton C, Bhattacharyya DR, Loughlin SO, Mohapatra PK, Mahanta J. Molecular characterization and species identification of the Anopheles dirus and An. minimus complexes in north-east India using r-DNA ITS-2 . Acta Trop. 2006;100(1–2):156–61. [DOI] [PubMed] [Google Scholar]
- 42. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences. 2012;109(16):6241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Le Goff G, Brengues C, Robert V. Stegomyia mosquitoes in Mayotte, taxonomic study and description of Stegomyia pia n. sp. Parasite. 2013;20:31 10.1051/parasite/2013030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Surendran S, Sarma D, Jude P, Kemppainen P, Kanthakumaran N, Gajapathy K, et al. Molecular characterization and identification of members of the Anopheles subpictus complex in Sri Lanka. Malar J. 2013;12(1):304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–9. [PubMed] [Google Scholar]
- 46. Crabtree MB, Savage HM, Miller BR. Development of a species-diagnostic polymerase chain reaction assay for the identification of Culex vectors of St. Louis encephalitis virus based on interspecies sequence variation in ribosomal DNA spacers. Am J Trop Med Hyg. 1995;53(1):105–9. [PubMed] [Google Scholar]
- 47. Brown JE, Evans BR, Zheng W, Obas V, Barrera-Martinez L, Egizi A, et al. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution. 2014;68(2):514–25. 10.1111/evo.12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. [DOI] [PubMed] [Google Scholar]
- 49. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Flot JF. Seqphase: A web tool for interconverting phase input/output files and fasta sequence alignments. Molecular Ecology Resources. 2010;10(1):162–6. 10.1111/j.1755-0998.2009.02732.x [DOI] [PubMed] [Google Scholar]
- 51. Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. [DOI] [PubMed] [Google Scholar]
- 55. Excoffier L, Lischer HE. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10(3):564–7. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 56. Maddison WP. Gene trees in species trees. Syst Biol. 1997;46(3):523–36. [Google Scholar]
- 57. Hlaing T, Tun-Lin W, Somboon P, Socheat D, Setha T, Min S, et al. Mitochondrial pseudogenes in the nuclear genome of Aedes aegypti mosquitoes: Implications for past and future population genetic studies. BMC Genetics. 2009;10:11 10.1186/1471-2156-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lyimo IN, Ferguson HM. Ecological and evolutionary determinants of host species choice in mosquito vectors. Trends Parasitol. 2009;25(4):189–96. 10.1016/j.pt.2009.01.005 [DOI] [PubMed] [Google Scholar]
- 59. Mutebi J-P, Barrett ADT. The epidemiology of yellow fever in Africa. Microb Infect. 2002;4(14):1459–68. [DOI] [PubMed] [Google Scholar]
- 60. Ellis BR, Sang RC, Horne KM, Higgs S, Wesson DM. Yellow fever virus susceptibility of two mosquito vectors from Kenya, East Africa. Trans R Soc Trop Med Hyg. 2012;106(6):387–9. 10.1016/j.trstmh.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 61. Sérié C, Andral L, Poirier A, Lindrec A, Neri P. Etudes sur la fièvre jaune en Ethiopie. 6. Etude épidémiologique. Bull W H O. 1968;38(6):879–84. [PMC free article] [PubMed] [Google Scholar]
- 62. Germain M, Monath P, Bryan J, Salaun JJ, Renaudet J. Yellow fever virus in the Gambia, 1978–1979: Entomological aspects and epidemiological correlations. Am J Trop Med Hyg. 1980;29(5):929–40. [DOI] [PubMed] [Google Scholar]
- 63. Lee VH, Moore DL. Vectors of the 1969 yellow fever epidemic on the Jos Plateau, Nigeria. Bull W H O. 1972;46(5):669–73. [PMC free article] [PubMed] [Google Scholar]
- 64. Port GR, Wilkes TJ. Aedes (Diceromyia) furcifer/taylori and a yellow fever outbreak in the Gambia. Trans R Soc Trop Med Hyg. 1979;73(3):341–4. [DOI] [PubMed] [Google Scholar]
- 65. Germain M, Cornet M, Mouchet J, Monath TP, Hervé J-P, Salaün JJ, et al. Recent advances in research regarding sylvatic yellow fever in West and Central Africa. Bulletin de L'Institut Pasteur. 1982;80(4):315–30. [Google Scholar]
- 66. Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85(2):328–45. 10.1016/j.antiviral.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, et al. Infection with Chikungunya virus in Italy: An outbreak in a temperate region. The Lancet. 2007;370(9602):1840–6. [DOI] [PubMed] [Google Scholar]
- 68. Thiboutot MM, Kannan S, Kawalekar OU, Shedlock DJ, Khan AS, Sarangan G, et al. Chikungunya: A potentially emerging epidemic? PLoS Negl Trop Dis. 2010;4(4):e623 10.1371/journal.pntd.0000623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pialoux G, Gaüzère B-A, Jauréguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. The Lancet Infectious Diseases. 2007;7(5):319–27. [DOI] [PubMed] [Google Scholar]
- 70. Josseran L, Paquet C, Zehgnoun A, Caillere N, Le Tertre A, Solet J-L, et al. Chikungunya disease outbreak, Reunion Island. Emerg Infect Dis. 2006;12(12):1994–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Van-Mai C-L, Claudine R, Anita T, Emilie R, Anne-Laure B, Henri-Pierre M, et al. Zika virus, French Polynesia, South Pacific, 2013. Emerging Infectious Disease Journal. 2014;20(6):1084. [Google Scholar]
- 72. Vireak H, Chadwick YY, Ly S, Andrew DH, Amelia PTdR, Robert BT, et al. Zika virus infection, Cambodia, 2010. Emerging Infectious Disease Journal. 2012;18(2):349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All sequence files are available from the GenBank database (accession number(s) KT998333- KT998452).