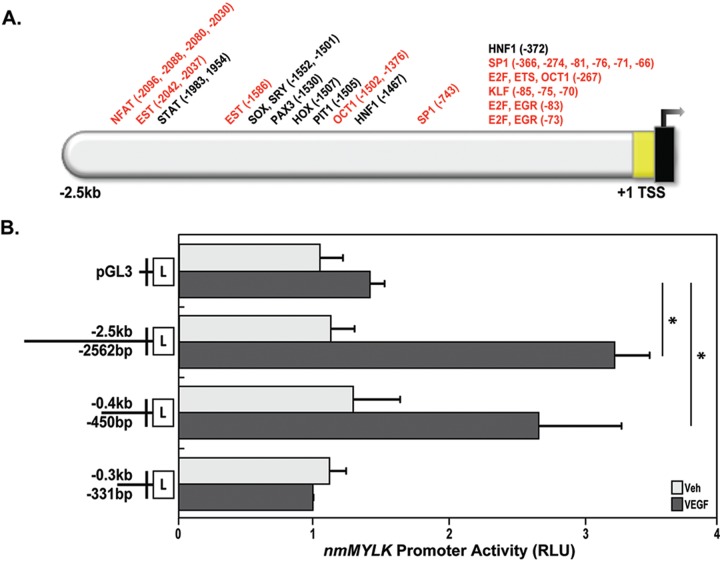
Figure 4.
nmMYLK promoter VEGF-associated transcription factor binding motif predictions and identification of a VEGF-responsive nmMYLK promoter region. A, Schematic depiction of predicted transcription factor binding sites (using Genomatix software) on the nmMYLK promoter (2.5 kb upstream of the transcription start site and noncoding exon 1). Thirty-six transcription factor binding modules are shown on top. Red indicates known VEGF effectors. B, Full-length nmMYLK promoter (−2.5 kb/−2,562 bp), nmMYLK promoter fragments (−0.4 kb/−450 bp, −0.3 kb/−331 bp), and pGL3 control luciferase constructs were overexpressed in lung endothelial cells and then treated with VEGF (100 ng/mL) for 24 hours. VEGF dramatically increases nmMYLK promoter activity in the 2.5-kb (full-length) and 0.4-kb fragments compared with that in the pGL3 control construct, as detected by luciferase reporter activity. However, the 0.3-kb fragment results in decreased luciferase activity compared with both the 2.5-kb (full-length) and the 0.4-kb fragment, indicating that the critical region for nmMLCK transcription by VEGF is between −0.4 kb/−450 bp and −0.3 kb/−331 bp on the nmMYLK promoter. Three independent experiments were performed, with bar graphs representing pooled promoter activity readings as relative light units (RLU). *P < 0.05 versus vehicle. TSS: transcription start site; VEGF: vascular endothelial growth factor; Veh: vehicle.