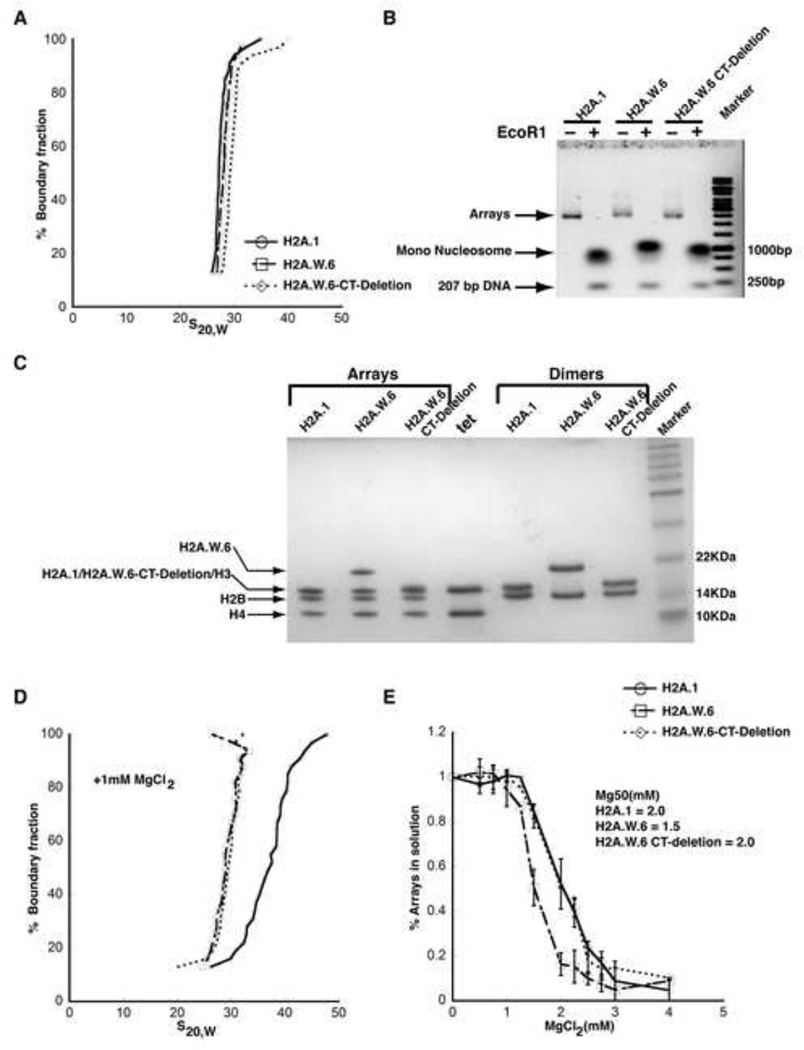
Figure 5. H2A.W causes chromatin condensation through chromatin fiber-fiber interactions promoted by its conserved C terminal tail in vitro.
(A) SV-AUC profile for 207-12 arrays assembled with H2A.1, H2A.W.6, and H2A.W.6-CT deletion indicates similar (and near-complete) saturation. (B) EcoRI digestion of nucleosome arrays to assay the degree of saturation of each array. Each array exhibits similar amounts of free 207 bp DNA confirming similar saturation of the arrays. (C) 1-2 µg of each array from (A) was run on a 15% SDS-PAGE to confirm equimolar distribution of histones. Tet indicates (H3-H4)2 tetramer. Note that H2A.1 and H2A.W.6-CT deletion runs in the same position as H3 on the gel. (D) The arrays were subjected to folding assay in the presence of 1 mM MgCl2. The H2A.1 containing control arrays show moderate folding (mid-point S ~40S), whereas both H2A.W.6 and H2A.W.6-CT deletion containing arrays do not undergo folding under these conditions. (E) Magnesium chloride-induced self-association assay showing that H2A.W.6 promotes inter-nucleosome array interactions. Arrays were incubated at the indicated concentrations of MgCl2, and the concentration of Mg2+ at which 50 % of the arrays have precipitated (Mg50) is listed in the figure (inset). Error bars are derived from two independent measurements (two preparations of reconstituted arrays).