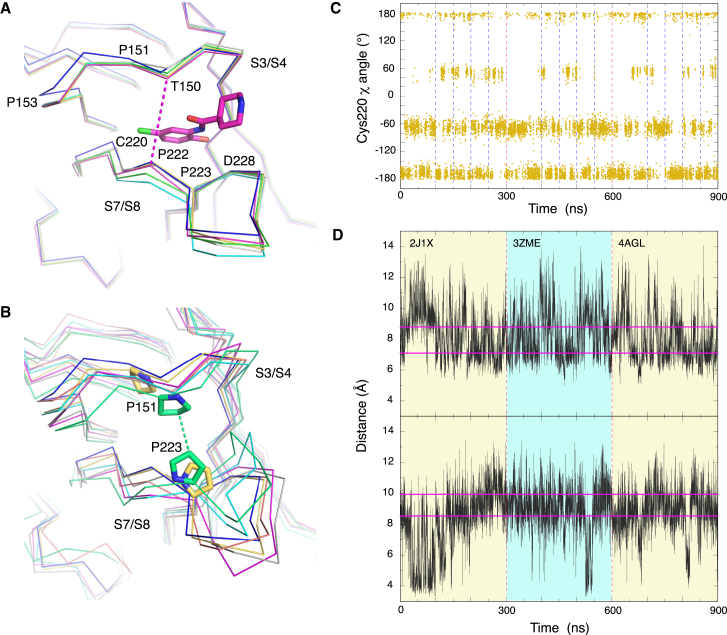
Figure 7.
Conformational Flexibility of the Y220C Pocket
(A) Superposition of the binding pocket in different crystal structures, showing shifts in the S7/S8 loop upon ligand binding that modulate the width of the binding pocket. Cα traces are shown for: ligand-free Y220C mutant (PDB: 2J1X) chains A (blue) and B (yellow), Y220C-12 complex chain B (magenta), Y220C-10 complex chain B (green), Y220C-7 complex chain B (cyan), and Y220C-1 complex chain B (gray; PDB: 2VUK). Bound molecule 12 is shown as a stick model. The distance between the Cα atoms of Thr150 and Pro222 in the Y220C-12 complex is highlighted with a magenta broken line.
(B) Superposition of representative structures from major clusters along the MD trajectory onto chains A (blue) and B (yellow) of the ligand-free crystal structure. The gray and magenta Cα traces are representative structures of the two largest clusters with an open pocket, comprising more than half of all structures along the trajectory. The green structure represents an ensemble of structures with a partial hydrophobic collapse of the pocket via interactions between Pro151 and Pro223. The close distance between the Cδ atoms of Pro151 and Pro223 in this structure is highlighted with a green broken line. The view is the same as in (A).
(C) Side-chain conformations of Cys220 (χ1 angle) along the MD trajectories. The conformation fluctuates between different rotamers corresponding to an open and close state of subsite III. Broken lines indicate starting points of different MD simulations, and those in red a new starting structure.
(D) Distance between the Cα atoms of Thr150 and Pro222 (top), and between the Cδ atoms of Pro151 and Pro223 (bottom) as a function of time over all simulations. The solid magenta lines indicate the largest and smallest distances observed in the various Y220C crystal structures. Starting and end points of the various simulations are the same as in (C).