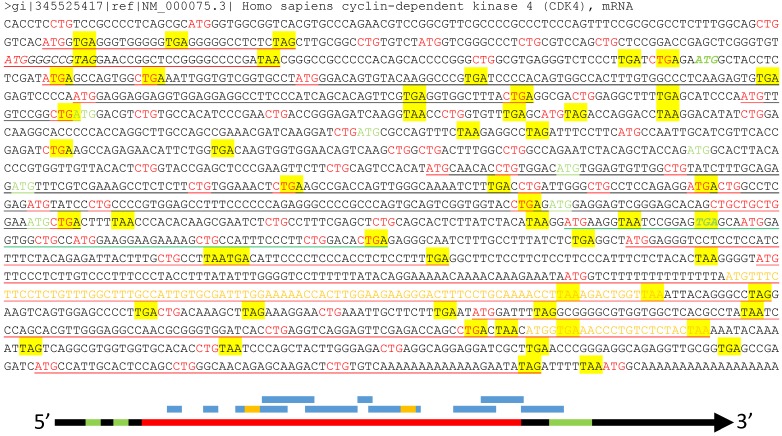
Fig 3.
Illustration of multiple ORFs in a given mRNA. Top panel: In the wt human CDK4 mRNA (copied from the NCBI database as a DNA sequence), as an example, all ATGs and CTGs as the most possible start codons are highlighted in red color while the three canonical stop codons (TAA, TAG and TGA) are shaded with yellow color. The ATG and TGA of the annotated CDK4 ORF are italicized and boldfaced with green color, while all in-frame downstream ATGs that may initiate N-terminally truncated CDK4 protein isoforms are highlighted in green color. Some (but not all, to avoid overwhelming the picture) ORFs that are initiated from out-of-frame ATGs and thus encode non-CDK4 peptides or proteins are underlined, with red underlining indicating the ORFs outside the CDK4 coding region, green underlining indicating an ORF overlapping with the CDK4 C-terminus, and black underlining indicating the ORFs within the CDK4 coding region. Some of these non-CDK4 AltORFs also contain some shorter out-of-frame AltORFs, which are displayed in yellow letters. Bottom panel: Although the current translation algorithm assigns only one ORF (long red bar, referred herein to as “annotated” ORF) to one mRNA (long black arrow), the mRNA also has two uORFs (short green bar) at the 5'UTR and an out-of-frame AltORF at the 3'UTR (long green bar). Moreover, there are many other short AltORFs (blue bar) that are not in frame with one another or with the annotated one. Some of these AltORFs may overlap with the nearby ones and contain some even shorter AltORFs (short yellow bar).