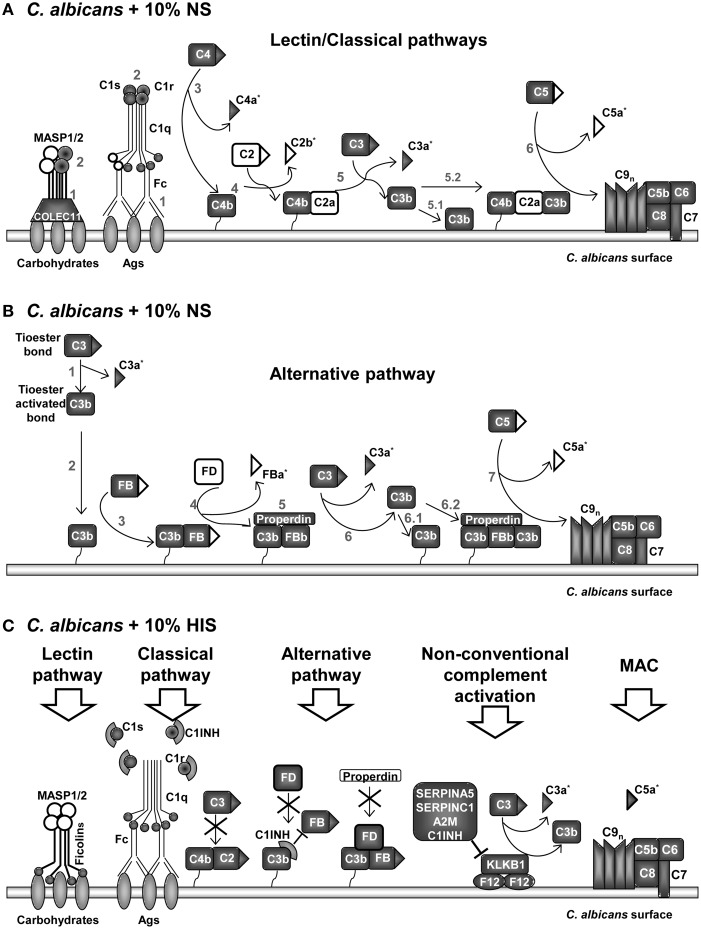
Figure 4.
Model of interactions among proteins of the complement pathways identified on C. albicans surface after incubation with human serum. Proteins identified in normal serum (NS) samples in (A,B). Proteins belonging to lectin and classical pathways are represented in (A) and to alternative pathway in (B). Proteins mainly identified on heat inactivated serum (HIS) in (C). Identified proteins on samples are indicated with gray background and not-identified in white. Complement fragments released during complement activation are indicated with triangles. Peptides belonging to complement fragments that can be released or be part of a full-length protein are indicated with *. Numbers in (A,B) correspond to sequential steps during complement pathways activation. (A) Collectin-11 was only identified in NS samples. MASP2 was identified in NS and MASP1 was not identified in any sample. Fc (constant region of Ig) of IgGs or IgMs start classical complement pathway. (B) Properdin was identified in NS and Factor D (FD) was not. (C) Ficolin-2, Ficolin-3, C1qA, C1INH, and FD were identified only in HIS samples. C1INH dissociates the C1qrs complex, blocks alternative C3 convertase and inhibits C3 activation by kallikrein. C2 was identified in HIS, including 4 peptides of C2b. Five peptides belonging to FBa were identified in HIS condition. Signally, peptides belonging to C5a fragment were only identified in HIS samples.