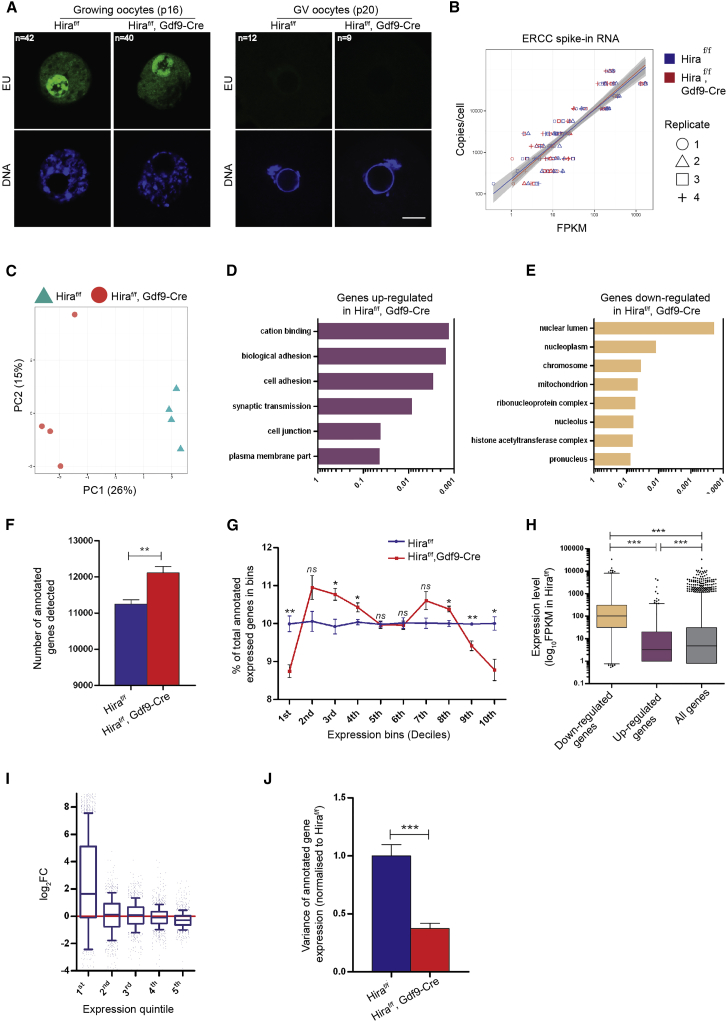
Figure 4.
Continuous Histone Replacement Mediated by Hira Is Required to Maintain the Full Dynamic Range of Gene Expression
(A) Detection of newly synthesized RNA by EU incorporation in growing (P14) and GV (P20) Hiraf/f and Hiraf/fGdf9-Cre+ oocytes is shown. Scale bar, 10 μm.
(B) Comparison between obtained FPKM values and absolute copy number per cell for ERCC spike-in RNA in Hiraf/f and Hiraf/fGdf9-Cre+ MII oocytes revealed no significant difference.
(C) Principal component analysis of scRNA-seq data derived from Hiraf/f and Hiraf/fGdf9-Cre+ MII oocytes is shown.
(D and E) Selected gene ontology (GO) terms significantly enriched for among differentially upregulated (D, edgeR, FDR < 0.1) and differentially downregulated (E, edgeR, FDR < 0.1) genes in Hiraf/fGdf9-Cre+ MII oocytes are shown. x axis represents Benjamini-Hochberg adjusted p value.
(F) Number of annotated genes detected, as computed by HTSeq program, is shown.
(G) Relative proportion of genes distributed among ten equally sized expression level bins. All genes detected in our RNA-seq experiment were divided into ten equal bins based on their expression levels in Hiraf/f oocytes. The gene number in each bin was counted for Hiraf/f or Hiraf/fGdf9-Cre+, and the percentage was calculated relative to all genes detected in a given sample.
(H) Boxplot shows gene expression levels of differentially upregulated (edgeR, FDR < 0.1), downregulated (edgeR, FDR < 0.1), and all annotated genes in Hiraf/fGdf9-Cre+ MII oocytes.
(I) Boxplots show distribution of gene expression fold change for each gene expression level quintile (based on Hiraf/f gene expression levels).
(J) Variance of gene expression within a given Hiraf/f or Hiraf/fGdf9-Cre+ sample. In all cases, error bars indicate SEM. Statistical analysis was carried out using two-tailed unpaired Student’s t test (F and G), Kruskal-Wallis with Dunn’s post hoc test (H) or F test (J); ns, non-significant; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. See also Figure S6.