Figure 5.
Hira Is Essential for Transcriptional Transitions Associated with the Oocyte Developmental Program and Is Required for Repression of Aberrant Transcription
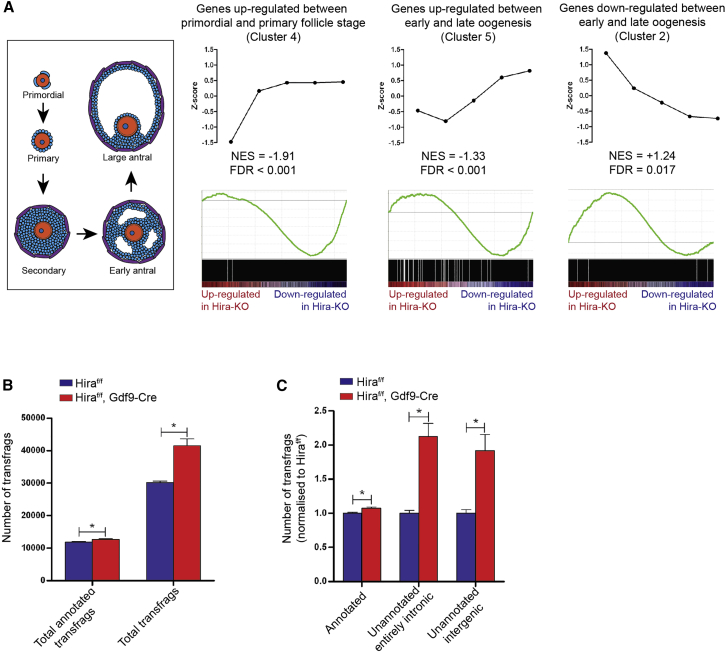
(A) GSEA comparing genes of selected expression clusters during oogenesis (Figure S5B) and the ranked list of gene expression changes in Hiraf/fGdf9-Cre+ MII oocytes relative to Hiraf/f oocytes. NES and FDR both were calculated in the GSEA program. For each cluster, each dot represents mean-normalized gene expression for consecutive stages of oocyte development.
(B) Numbers of total transfrags and total annotated transfrags in Hiraf/f and Hiraf/fGdf9-Cre+ MII oocytes (as computed by CuffCompare program) are shown.
(C) Numbers of total annotated transfrags, unannotated entirely intronic transfrags, and unannotated entirely intergenic transfrags in Hiraf/f and Hiraf/fGdf9-Cre+ MII oocytes (as computed by CuffCompare program) are shown. In all cases, error bars indicate SEM. Statistical analysis was carried out using two-tailed Student’s t test (∗p < 0.05). See also Figures S5 and S6.