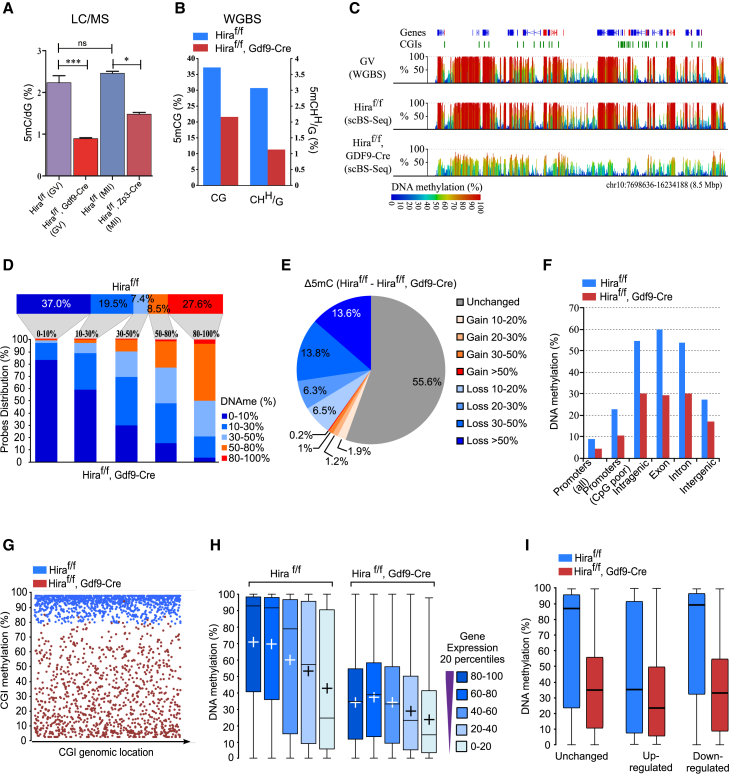
Figure 6.
Continuous Histone Replacement Is Required for Efficient De Novo Methylation during Oogenesis
(A) Reduced total 5mC was measured by LC-MS in GV (Hiraf/f and Hiraf/fGdf9-Cre+) and MII (Hiraf/f and Hiraf/fZp3-Cre+) oocytes.
(B) Global levels of DNA methylation quantified by scBS-seq in the CpG (CG) and non-CpG (CHH/G) contexts are shown.
(C) Example shows CpG methylation quantified over 3-kb sliding windows (1.5-kb steps) for published GV datasets (Shirane et al., 2013; WGBS, top), Hiraf/f, and Hiraf/fGdf9-Cre+ (scBS-seq).
(D) Distribution shows the 3-kb genomic windows in the indicated bins of DNA methylation in Hiraf/f oocytes (top, horizontal, percentage indicating the proportion of methylation bins) and their corresponding DNA methylation values in Hiraf/fGdf9-Cre+ oocytes (bottom, vertical columns).
(E) Pie chart distribution shows the 3-kb genomic windows presenting statistically significant (chi-square test, p < 0.01) changes in Hiraf/fGdf9-Cre+ versus Hiraf/f (percentage indicates the proportion of each segment).
(F) Effect of Hira deletion on DNA methylation is global and independent of the genomic context. DNA methylation at CpGs (five reads coverage) was determined and averaged for each genomic context.
(G) CpG islands (CGIs) methylated in Hiraf/f oocytes are globally hypomethylated in Hiraf/fGdf9-Cre+ oocytes. CGI methylation was defined for each genotype, and only CGIs hypermethylated in Hiraf/f (>80%) are displayed, for both Hiraf/f (blue) and Hiraf/fGdf9-Cre+ (red), and ordered on the x axis based on their genomic location.
(H) Effects on DNA methylation in Hiraf/fGdf9-Cre+ oocytes are more pronounced at highly expressed genes. DNA methylation was quantified for genes binned into expression percentile based on the scRNA-seq data (boxplot with plus signs representing mean values and horizontal bars representing median values).
(I) Comparison between DNA methylation and gene expression differences in Hiraf/f and Hiraf/fGdf9-Cre+ oocytes. See also Figure S7.