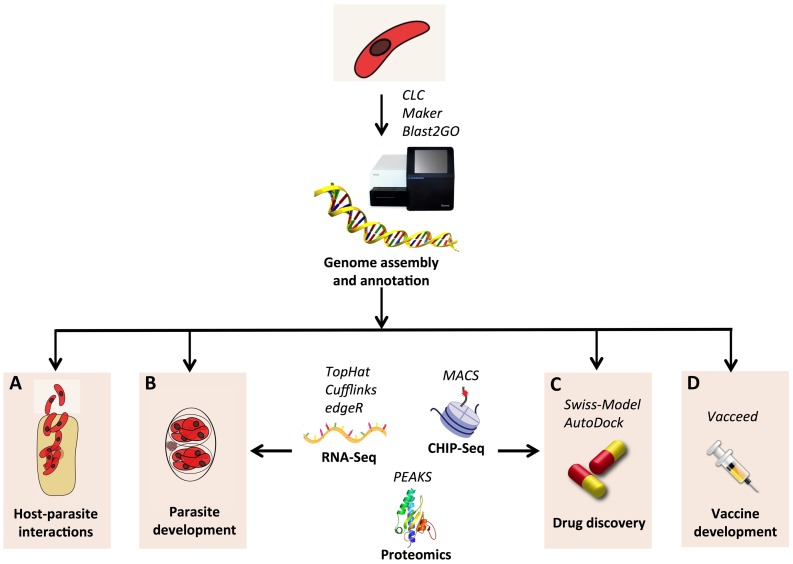
Figure 2.
Schematic view of the in silico analysis of genomic data for C. suis. The genome sequence can be assembled with next generation sequencing using a combination of short and long reads libraries. Tools such as CLC (CLC Bio-Qiagen, Aarhus, Denmark), Maker (88), and Blast2GO (70) can be applied for de novo assembly, annotation, and functional annotation, respectively. Other NGS technologies, such as RNA-Seq (89) and CHIP-Seq (90), together with proteomics (91), can be used to unravel the biology of the parasite and to discover new drugs and vaccines. Changes involved in host–parasite interaction (A) and developmental switches (B) can be identified both at the genetic and epigenetic level by RNA-Seq and CHIP-Seq, respectively: transcripts are reconstructed using the programs TopHat (92) and Cufflinks (93); differentially expressed genes are detected by edgeR (94); CHIP-Seq data are processed with the MACS software (95). (C) 3D structure of drug candidates can be reconstructed by homology using Swiss-Model (96); screening of virtual libraries of compounds can be performed with AutoDock (97). (D) Vaccine candidates can be identified using Vacceed (86) and validated by proteomics approaches, such as mass spectrometry, with the aid of the software PEAKS (Bioinformatics Solutions Inc., Waterloo, ON, Canada).