Figure 2. Multiplex detection and identification of viruses.
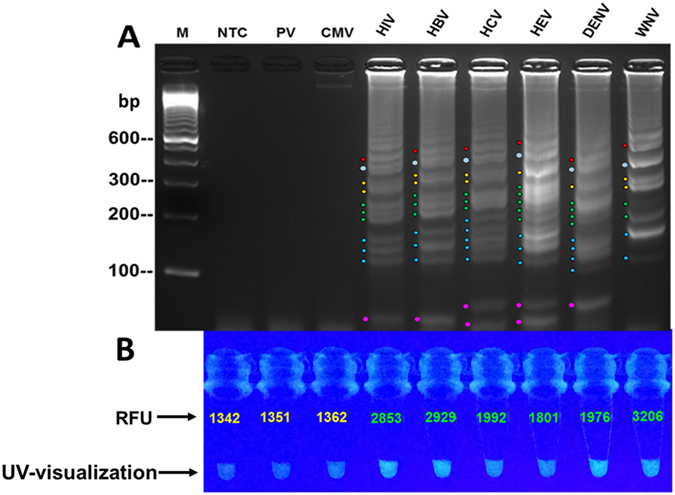
Standard reference viral nucleic acids were subjected to isothermal multiplex amplification to evaluate the assay’s detection ability utilizing virus-specific primers-sets and specific fluorogenic oligonucleotides. Detected targets were identified by banding-pattern analysis. Targets were amplified with a master reaction mixture containing all the primer-sets and all the 6-FAM/BHQ1-tagged fluorogenic oligonucleotides. Differences in banding patterns were observed and identified by the location of pattern formation in relation to the molecular marker, size of the bands within the patterns, spacing of each group of patterns and the laddering-shift of the patterns (also see Supplementary Figures S1-S4). (A) Electrophoretic analysis showed detection of all target nucleic acids with distinguishing ladder-like banding patterns in lanes 4–9 for human Immunodeficiency virus (HIV), hepatitis-B virus (HBV), hepatitis-C virus (HCV), hepatitis-E virus (HEV), dengue virus (DENV), and West Nile (WNV). No detection was observed in the negative controls which includes the non-template control (NTC in lane 1), cytomegalovirus (CMV in lane 2), and, parvovirus (PV in lane 3); M = 100 bp marker. Note that for WNV distinct pattern generation, W-LF may not be required in reaction. (B) Fluorimetric investigation revealed positive fluorescence detection of HIV, HCV, HBV, HEV, DENV, and WNV as demonstrated by high fluorescence intensity and higher relative fluorescence unit (RFU) as compared with the NTC, CMV, and PV controls.
