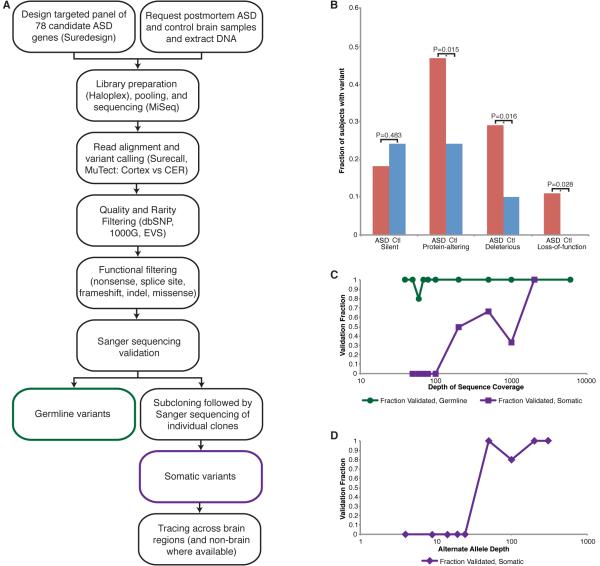
Figure 1. Targeted deep sequencing identifies germline and somatic mutations in ASD candidate genes in postmortem ASD brain.
A. Study schematic. DNA extracted from ASD and control brain samples was sequenced at high coverage using a targeted panel of candidate ASD genes. After variant calling and filtering, protein-altering variants were validated using Sanger sequencing and subcloning, and somatic variants were traced across the brain and non-brain tissues, when available. B. Fraction of cases and controls harboring synonymous, protein-altering, deleterious (subset of protein-altering), or loss-of-function (subset of deleterious) variants. p values were calculated using a two-tailed Fisher’s exact test. C. Validation of germline and somatic variants according to total depth of sequence coverage. Essentially all germline variants validated; somatic variants were more likely to validate at higher total depth (≥100X). D. Validation of somatic variants according to alternate allele depth of coverage. Somatic variants were more likely to validate at alternate allele depth ≥25X. Each data point represents the validation fraction for all variants with b. total or c. alternate allele depth from the previous data point to that data point. See also Tables S1, S3-S5.