Figure.
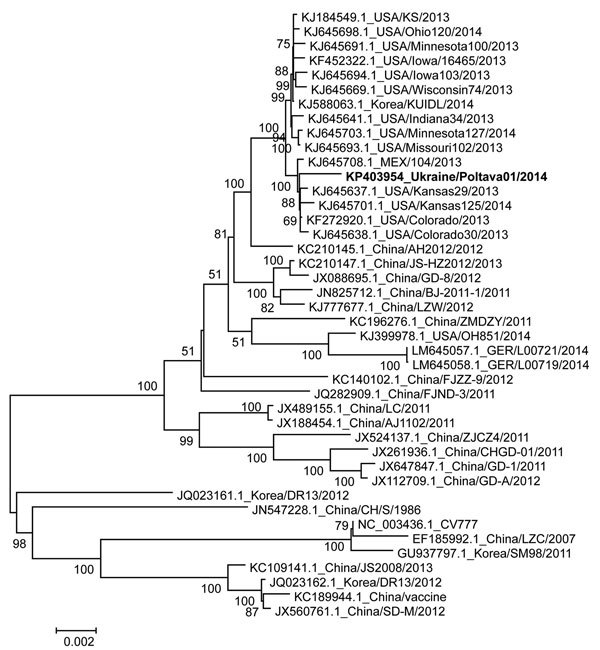
Phylogenetic analysis of the full-length genome of the porcine epidemic diarrhea virus (PEDV) Ukraine/Poltava01/2014 (bold text). The full-length genomes of PEDV were aligned by using the MegAlign software of the DNASTAR Lasergene Core Suite (DNASTAR, Inc., Madison, WI, USA) and phylogenetic analysis was done by using MEGA 5.2 software (13). The tree was constructed by using the neighbor-joining method and 1,000 bootstrap replications. Only bootstrap values of more than 50% are shown in the figure. Each virus on the tree is represented by accession number, strain, and year of sample collection. Ukraine/Poltava01/2014 clusters in close proximity to recent strains in the United States other than the Ohio851 variant, and both are substantially genetically different from the previous European variants, such as the prototype strain CV777, which is embedded in another cluster. Scale bar indicates nucleotide substitutions per site.
