Figure 2.
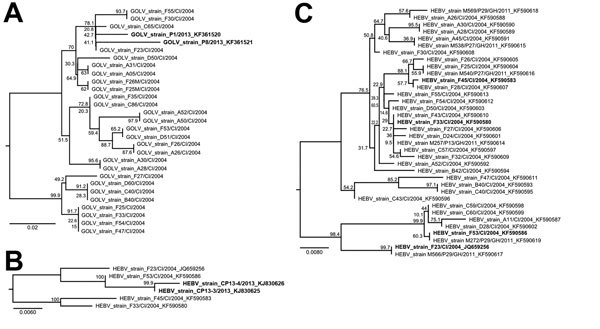
Maximum-likelihood phylogenetic analyses of Gouléako virus (GOLV) and Herbert virus (HEBV) strains from mosquitoes in Côte d’Ivoire, 2004, and Ghana, 2011, and virus strains detected by Chung el al. (9) in pigs in South Korea. A) Analysis of the glycoprotein precursor gene of GOLV strains identified in mosquitoes collected in Côte d’Ivoire and Ghana and of strains detected in swine in South Korea. Sequences originating from swine are shown in bold. B) Analysis of the RNA-dependent RNA polymerase gene of HEBV strains from mosquitoes and swine. Sequences originating from swine are shown in bold. C) Analysis of all identified HEBV strains found in mosquitoes. HEBV strains used for phylogenetic analyses in panel B are shown in bold. GOLV strains F25M/CI/2004 and F26M/CI/2004 were found in male mosquitoes. Scale bars indicate nucleotide substitutions per position in the alignment.
