Fig. 1. PRMT8 knockout mice display abnormal Purkinje cell dendrites and aberrant behavior.
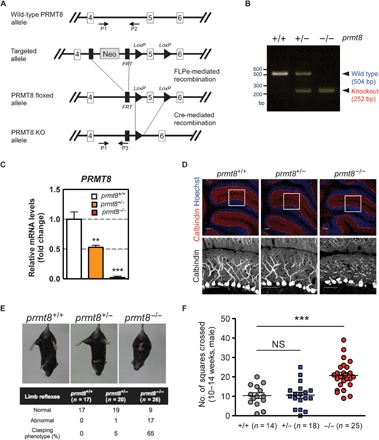
(A) Gene targeting strategy for the prmt8 locus: schematic representation of the prmt8+/+ [wild-type (WT)], prmt8flox (floxed), and prmt8−/− (knockout) alleles. Arrows indicate the positions of the P1, P2, and P3 genotyping primers. (B) Allele-specific PCR analysis using tail genomic DNA. Products of 504 and 252 base pairs (bp) were generated from prmt8+/+ and prmt8−/− mice, respectively. (C) prmt8 mRNA levels in 12-week-old mice were examined in total cerebellar RNA by qPCR. Results are shown as fold change of prmt8 mRNA expression relative to the mean WT value. Data were obtained using the ΔΔCt method with normalization to the reference glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (mean ± SEM, n = 4 per genotype; **P < 0.05, ***P < 0.0001). (D) Top: Representative confocal images of cerebellar sections from 12- to 14-week-old animals (genotyped as indicated) stained for immunofluorescence detection of calbindin (red) to indicate Purkinje cells and Hoechst 33258 (blue) to label DNA (scale bars, 20 μm). Bottom: Enlarged images of the boxed areas with the Purkinje cell layer (scale bars, 20 μm). (E) Limb-clasping reflex in 10- to 14-week-old mice suspended by the tail as monitored by video recording. The percentage indicates the limb-clasping reflex of mice (genotyped as indicated). (F) Locomotor activity of prmt8−/− mice. The total number of squares crossed by the insect during 30 s is indicated. Data are shown as means ± SEM; ***P < 0.0001 or NS (not significant) as compared to WT animals.
