Fig. 1.
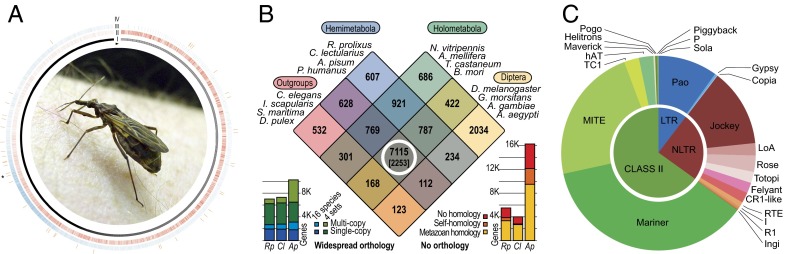
Rhodnius genome. (A) Genome overview. All scaffolds are represented in layer I and are organized clockwise from the longest to the shortest, starting at the arrowhead. The genic (layer II, red) and TEs (layer III, blue) showed opposite densities until the asterisk (*) and were similarly low from this point until the end (shorter scaffolds). The Wolbachia sp. insertions (layer IV, orange) were observed throughout the genome without a trend. The kernel picture illustrates an adult R. prolixus. (B) Gene clustering. The Venn diagram partitions 15,439 OrthoMCL gene clusters according to their species compositions for R. prolixus and three other Hemimetabola (blue), four Diptera (yellow), four other Holometabola (green), and four noninsect outgroup species (pink). The 6,993 R. prolixus genes show widespread orthology (white circle, and bars, Bottom Left): these are part of the 7,115 clusters that have representatives from each of the four species sets, of which a conserved core of 2,253 clusters have orthologs in all 16 species. The 5,498 R. prolixus genes show no confident orthology (bars, Bottom Right), but most of these are homologous (e-value < 1e-05) to genes from other animals or to genes in its own genome. (C) TE distribution. The inner chart represents the three main classes of TEs (LTRs, non-LTRs, and class II), and the outer shows the distribution of TE superfamilies within each class. The charts are based on the total base pairs occupied by TE-related sequences longer than 0.5 Kb, as shown in SI Appendix, Table A1.