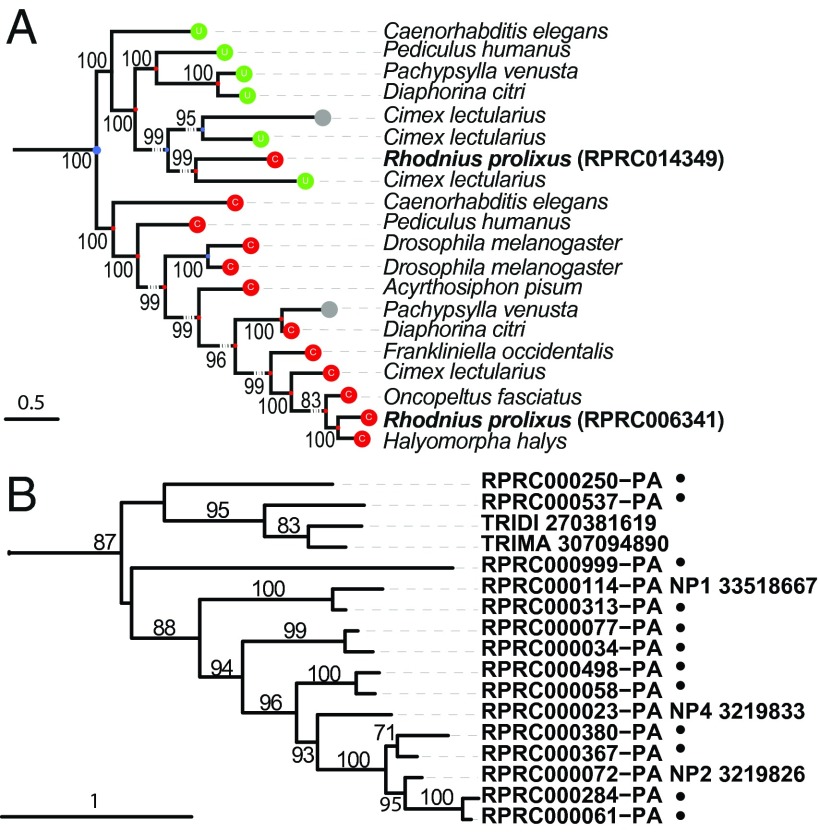
Fig. 3.
Thioredoxin reductases and nitrophorins trees. (A) The tree shows the phylogenetic relationship of thioredoxin reductases (TR) in nine Paraneoptera genomes. Colored balls indicate the amino acid aligned to the Sec position: green (U) selenocysteine, red (C) cysteine, and gray (–) unknown/unaligned. One of the two Cys-TR in R. prolixus (RPRC014349) clusters with the Sec-TR proteins, implying a Sec to Cys conversion. (B) Nitrophorins branch of lipocalins tree exemplify a Rhodnius enriched cluster. Sequences clustered by OrthoMCL on group 691 together with unclustered sequences similar to lipocalins were aligned and a tree constructed (SI Appendix, Fig. A15). Black dots mark new Rhodnius sequences. TRIDI, Triatoma dimidiata; TRIMA, Triatoma matogrossensis. Support values based on bootstrap are included at nodes.