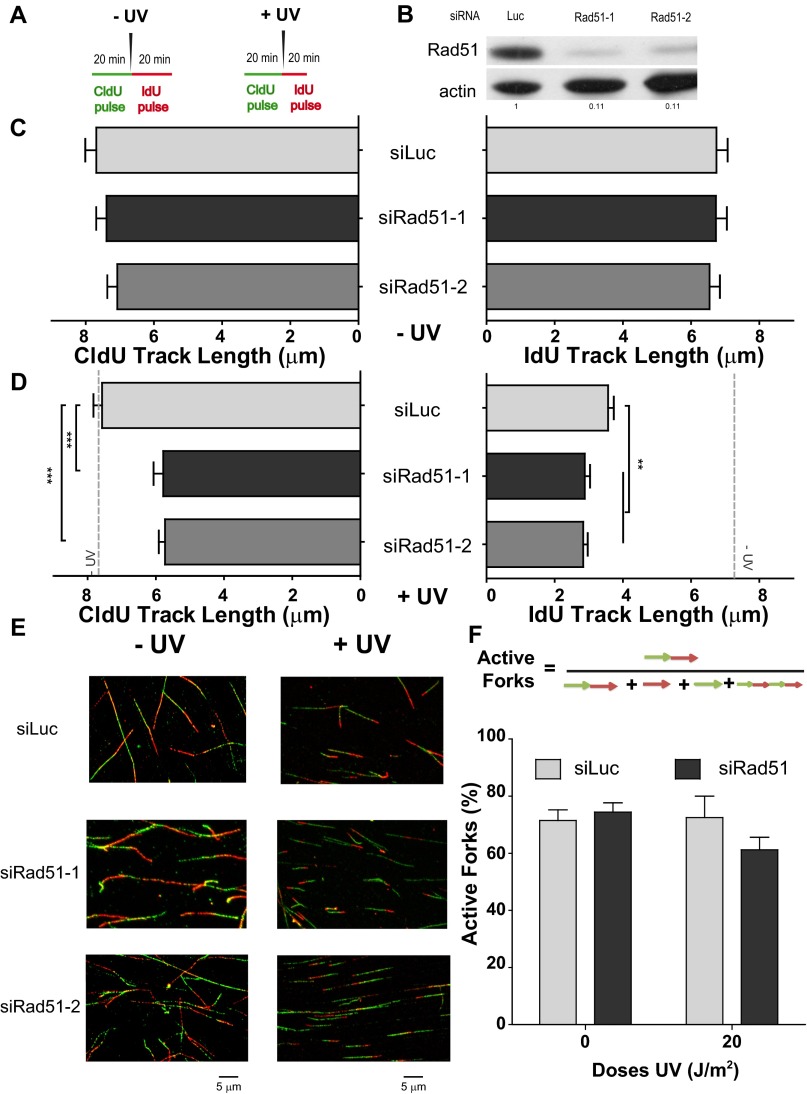
Fig. S3.
Both the CldU and IdU track lengths are modified when Rad51-depleted cells are UV-irradiated (U2OS cells). (A) Schematic of the DNA fiber-labeling experiment. (B) Western blot analysis of U20S cells treated with the two independent siRNAs for Rad51 (siRad51-1 corresponds to sequence 1 and is the siRNA used throughout the study, and siRad51-2 corresponds to sequence 2). Antibodies specific for Rad51 and actin were used. (C) CldU (Left) and IdU (Right) track length quantification in sham-irradiated samples. (D) CldU (Left) and IdU (Right) track length quantification of UV-irradiated samples. For reference, the average CldU and IdU track lengths of the control sham-irradiated conditions are shown as a gray dotted line. (E) Representative panels corresponding to experiments shown in C and D. (F) Quantification of active forks visualized as bicolor fibers (red and green contiguous tracks). Percentages of the total number of fibers scored, including origins (only IdU label), terminations (only CldU label), and interspersed (multiple CldU and IdU labels), are shown. In all quantifications, mean values and SDs are shown. Two independent experiments were analyzed obtaining similar results. For all figures in this study, the significance of the differences is as follows: *P < 0.1; **P < 0.01; ***P < 0.001. If the P value is not indicated, the difference is not statistically significant. Error bars represent SEM (SEM value).