Figure 1.
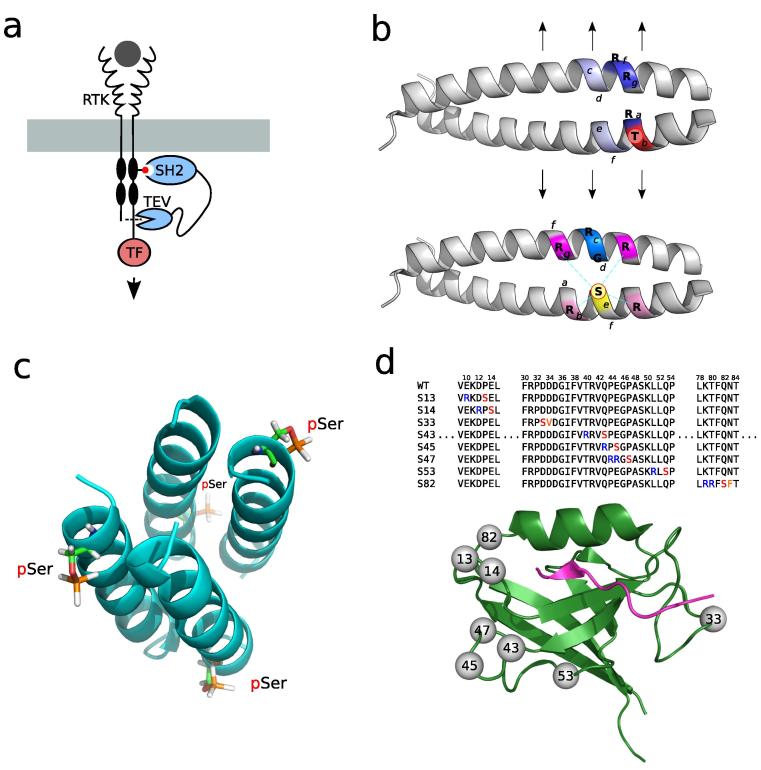
Design of post-translational regulation of protein–protein interactions. (a) The binding of phosphorylation ‘reader’ domains, like the SH2 domain, to their target peptides has been well characterized for some proteins. These rules can be used to engineer novel PTM regulated interactions like the recruitment of the TEV protease to an active RTK. Activation of the receptor causes the phosphorylation of specific tyrosine residues that recruit the binding of the SH2 domain. The recruited TEV can cleave off a transcription factor causing a downstream gene-expression response. (b) The leucine zipper pair illustrated here is a model for studying dimerization. Szilák and colleagues used this model system to study how the introduction of phosphorylation sites at different positions could regulate this protein interaction. The lower-case letters indicate the positions along the helix. In the example described in the upper diagram the phosphosite was introduced at a position ‘b’ (coloured red) that points way from the interface but causes a destabilization of the helix (at the positions coloured blue) and therefore inhibit the dimmer formation (arrows). The lower diagram illustrates the addition of a phosphosite at position ‘e’ (coloured yellow) pointing towards the interface where the phosphosite can interact with two opposing arginines (coloured magenta) and stabilize the interaction. (c) The Lac repressor oligomerization domain is another model system used to study protein–protein interactions. Signarvic and DeGrado found that phosphosites introduced in the monomer towards the N-terminus, illustrated here, caused an increase in the stability of the tetramer. (d) The structure of a Erbin PDZ (PDB: 1MFG) bound to a target peptide is shown. The positions highlighted were selected for the introduction of an engineered target site for the PKA kinase. The corresponding mutations needed to create a PKA target site are shown in the sequences above the structure.