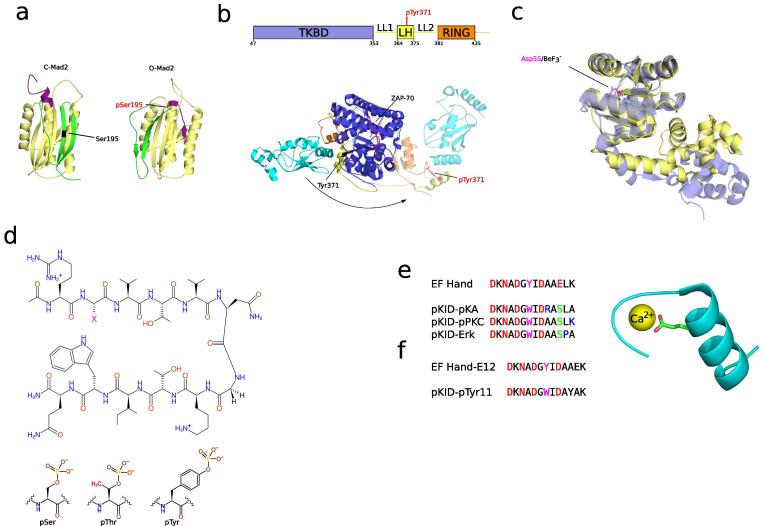
Figure 2.
Allosteric regulation by post-translational modifications. (a) The structures of the open (O-Mad2, PDB: 1DUJ) and closed (C-Mad2, (PDB: 1S2H) conformations of the spindle checkpoint protein Mad2 are shown. The phosphorylation of serine 195 promotes the open conformation. The protein segments coloured in yellow show an overall similar conformation. Sections in green indicate the regions showing the highest change in conformation. The N-terminal region is highlighted in purple in both conformations. (b) The RING ubiquitin ligase c-Cbl contains 3 globular domains connected by linkers illustrated in the upper diagram. This protein can adopt two different conformations that are regulated by protein phosphorylation. In the unphosphorylated form the LH domain packs against the RING and TKBD domain (opaque structures, PDB: 1FBV). After phosphorylation LH and RING domain change their orientation relative to the TKBD domain drastically towards the binding interface of the target protein (PDB: 4A4C). This changes the orientation and distance between the E2 and the target protein. (c) The unphosphorylated (yellow, PDB: 4GVP) and beryllofluoride-actived (purple, PDB: 4IF4) conformations of the two-component response regulator VraR. Beryllofluoride (BeF3−) is mimicking phosphorylation. (d) Structure of a 12 residue peptide β-hairpin used to study the consequence of phosphorylation on allosteric regulation. The second position (X in the figure) of the hairpin was changed to different phosphosites and the impact on hairpin formation was evaluated. (e) and (f) The EF-hand helix-loop-helix structure was used to design a serine (e) and tyrosine (f) kinase sensor by introducing the phosphorylation sites highlighted green in the sequences. Evolutionary conserved glutamate at position 12 is highlighted in the sequences and structure.