Fig. 1.
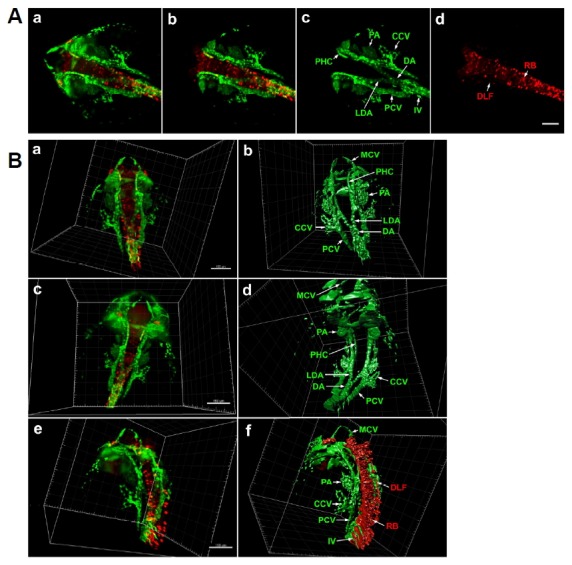
Light-sheet microscopy of differentiating neurons and vasculature in Tg(elavl3: DsRed/fli1a: EGFP) zebrafish embryo at 24 hpf. (A) Dorsal view of whole head and anterior trunk. (a) 3D reconstruction of the entire anterior portion of embryo. fli1a: EGFP-positive cells are green and elavl3:DsRed-positive cells are red. Data is comprised of 1740 optical sections of 0.488 μm thickness. (b) 3D reconstruction of 740 central optical sections (1001–1740), omission of dorsal and ventral regions allows clearer visualization of internal fli1a:EGFP-positive (c) and elavl3: DsRed-positive (d) structures. Scale bar 100 μm. (B) 3D reconstruction and segmentation using Imaris software. (a) 3D reconstruction of full image stack in dorsal view. (b) 3D segmentation of fli1a: EGFP-positive tissues, internal structures are labelled. (c) Ventral view of same embryo. (d) 3D segmentation of fli1a:EGFP-positive tissues. (e) Lateral view of 24 hpf embryo. (f) 3D segmentation of fli1a:EGFP and elavl3:DsRed-positive tissues. An animated 3D video of these data is presented in Supplementary Fig. S1. Scale bar 150 μm. CCV, common cardinal vein; DA, dorsal aorta; DLF, dorsal longitudinal fasciculus; IV, intersegmental vein; LDA, lateral dorsal aorta; MCV, middle cerebral vein; PA, pharyngeal arch; PCV, posterior cardinal vein; PHC, primordial hindbrain channel; RB, Rohon-Beard neuron.
