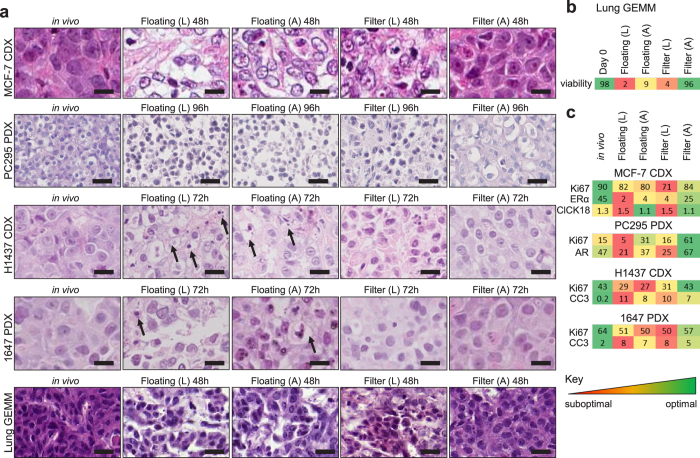
Figure 2. Histopathology of cultivated tumour slices.
(a) High power images of H&E stained tumour tissue slice sections, representing each tumour model cultivated under different conditions, compared to a stained section of the original in vivo tumour (CDX = cell line-derived xenograft; PDX = patient-derived tumour; GEMM = genetically engineered mouse model). Rows H1437 CDX and 1647 PDX: arrows indicate condensed nuclei (apoptosis) and vaculated regions. Representative images from at least three experiments are shown. Scale bars represent 25 μm. (b) Heatmap of the percentage of viable tissue in NSCLC GEMM tumour slices determined by the algorithm described in the Methods. (c) Heatmaps representing the percentage of positive cells expressing viability markers by immunohistochemistry (IHC) of slices cultured under different conditions (numbers in heatmap boxes represent the percentage of positive cells for each biomarker). Key to conditions: A – atmospheric oxygen, L – low oxygen. Ki67, indicating proliferation; ClCK18, cleaved cytokeratin 18 indicating apoptosis; ERα, estrogen receptor-alpha; AR, androgen receptor; CC3, cleaved caspase 3. The values reflect means from three independent experiments.