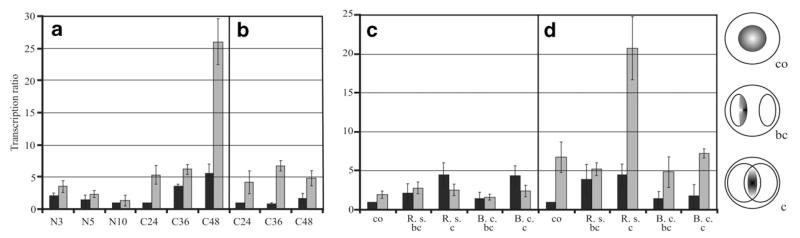
Fig. 3.
Relative transcription ratios of the chitinase-encoding genes nag1 (a and c) and ech42 (b and d) in the Δtmk1-11 mutant (grey bars) compared to the parental strain T. atroviride P1 (black bars). (a and b) Real-time RT-PCR analyses were performed 3, 5, and 10 h after transfer of mycelia to liquid growth medium containing 1% N-acetylglucosamine (N) and 36 and 48 h after transfer to 1% colloidal chitin (C). The samples of the parental strain showing the lowest mRNA levels within one PCR run were arbitrarily assigned the factor 1 (N10, C24). (c and d) Direct confrontation assays with R. solani (R.s.) and B. cinerea (B.c.) as host fungi. Samples were collected from a control (co) where T. atroviride was grown alone, from an early stage before direct contact between the two fungi (bc), and from a later stage of direct contact—if present—with an interaction zone of 1.0 cm (c) and subjected to real-time RT-PCR. The control sample of the parental strain was arbitrarily assigned the factor 1.